Identifying thousands of RNA genes in Brassicaceae
Palos et al. identify thousands of new long intergenic noncoding RNAs using public data.
By Kyle Palos and Andrew Nelson
Background: All plants have thousands of genes in their genomes that contribute to plant form and function. While the functional “end-state” of many of these genes are proteins, some genes produce RNAs that never produce a protein, but instead function as an RNA molecule. These non-protein coding RNAs (ncRNAs) are typically separated into two classes based on length: small ncRNAs and long ncRNAs (lncRNAs). Due to their low abundance, poor sequence conservation, and lack of obvious functional domains, lncRNAs are less studied compared to protein-coding genes. Despite these difficulties, some lncRNAs have been shown to be critical in regulating how plants grow and respond to changes in their environment.
Question: How many lncRNAs are present in plants, and what are their characteristics and functional roles? To start answering these questions, we examined thousands of RNA sequencing datasets from four mustards, including the model organism Arabidopsis thaliana (thale cress), a relative with high seed-oil content, Camelina sativa, the species that gives us bok choy and turnips, Brassica rapa, and the salt tolerant mustard Eutrema salsugineum.
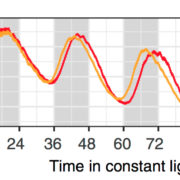
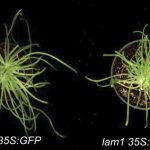
Findings: We found evidence for thousands of lncRNAs in each of the four species. These lncRNAs are often very tissue or context (stress) specific. Of the identified lncRNAs, we highlighted those that were unusually conserved or contained elements that might contribute to function. We also proposed functions for some lncRNAs based on patterns of abundance across tissues/conditions. Using this approach, we uncovered a set of lncRNAs that appear to be important for seed germination in Arabidopsis.
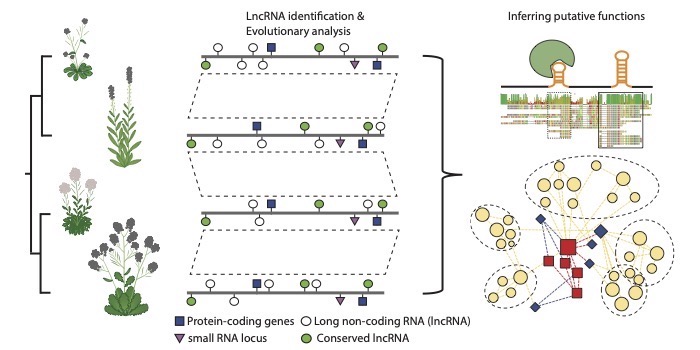 Next steps: Using the lncRNA resources generated in this project, we are examining the functions of those that we believe are critical for germination or responses to environmental stresses. In addition, we are expanding our identification efforts to other systems, including agriculturally significant species within the grasses.
Next steps: Using the lncRNA resources generated in this project, we are examining the functions of those that we believe are critical for germination or responses to environmental stresses. In addition, we are expanding our identification efforts to other systems, including agriculturally significant species within the grasses.
Kyle Palos, Anna Nelson Dittrich, Li’ang Yu, Jordan Brock, Caylyn Railey, Hsin-Yen Wu, Ewelina Sokolowska, Aleksandra Skirycz, Polly Hsu, Brian Gregory, Eric Lyons, Mark Beilstein, and Andrew Nelson (2022). Identification and Functional Annotation of Long Intergenic Non-coding RNAs in Brassicaceae. https://doi.org/10.1093/plcell/koac166