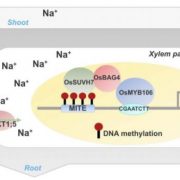
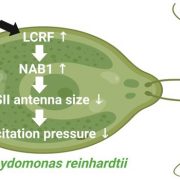
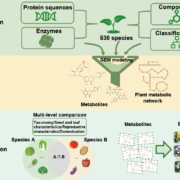
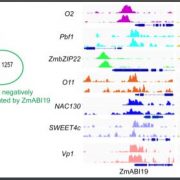
Identification of plant transcriptional activation domains
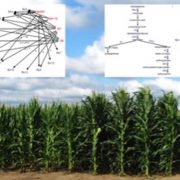
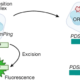
 Activation domains (AD) are parts of transcription factor proteins (TFs) that bind the transcriptional machinery (coactivator complexes) and lead to transcriptional activation. However, identifying ADs is challenging because they are often part of intrinsically disordered regions, which lack a defined 3D structure and conserved sequence motifs. In this paper, Morffy and colleagues adressed this challenge by combining large scale high throughput assays in yeast with the implementation of a neural network to identify and classify the ADs of plant TFs. To test the activity of ADs and identify their sequences, they created a library of 40aa long peptides spanning the whole coding sequence of 1918 TFs from Arabidopsis, and employed it in a fluorescence-based reporter system followed by sequencing. The experimental data were used to train a neural network that could identify ADs based on a set of predefined biochemical features derived from the amino acidic sequences. By extracting the features with the strongest effects from the model, the authors classified the ADs into six different subtypes, whose activity was then verified in planta. Finally, the authors applied the neuronal network to members of the Auxin Response Factor (ARFs) family across 117 angiosperms. They found that while the sequence similarity of ADs was low among orthologs, their location within the protein was conserved. This work provides unprecedented insights on the physico-chemical properties of TFs and, more broadly, on the mechanisms governing transcriptional activation. It emphasizes the role of positional and functional conservation, rather than sequence homology, in the evolution of TFs. (Summary by Carlo Pasini @Crl_Psn) Nature 10.1038/s41586-024-07707-3
Activation domains (AD) are parts of transcription factor proteins (TFs) that bind the transcriptional machinery (coactivator complexes) and lead to transcriptional activation. However, identifying ADs is challenging because they are often part of intrinsically disordered regions, which lack a defined 3D structure and conserved sequence motifs. In this paper, Morffy and colleagues adressed this challenge by combining large scale high throughput assays in yeast with the implementation of a neural network to identify and classify the ADs of plant TFs. To test the activity of ADs and identify their sequences, they created a library of 40aa long peptides spanning the whole coding sequence of 1918 TFs from Arabidopsis, and employed it in a fluorescence-based reporter system followed by sequencing. The experimental data were used to train a neural network that could identify ADs based on a set of predefined biochemical features derived from the amino acidic sequences. By extracting the features with the strongest effects from the model, the authors classified the ADs into six different subtypes, whose activity was then verified in planta. Finally, the authors applied the neuronal network to members of the Auxin Response Factor (ARFs) family across 117 angiosperms. They found that while the sequence similarity of ADs was low among orthologs, their location within the protein was conserved. This work provides unprecedented insights on the physico-chemical properties of TFs and, more broadly, on the mechanisms governing transcriptional activation. It emphasizes the role of positional and functional conservation, rather than sequence homology, in the evolution of TFs. (Summary by Carlo Pasini @Crl_Psn) Nature 10.1038/s41586-024-07707-3