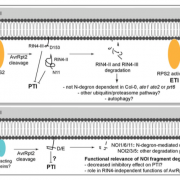
Highly active rubiscos discovered by systematic interrogation of natural sequence diversity (EMBO J)
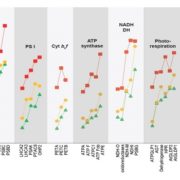
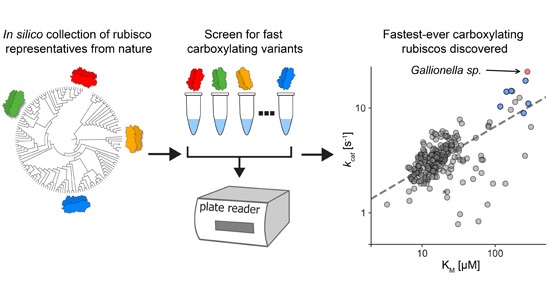 This is a fascinating and very well-written paper that investigates the diversity of rubisco’s kinetic properties. Rubisco’s relationship with its substrate CO2 is complicated by its relationship with O2, and it has often been suggested that for this reason rubisco is locked into a slow rate of catalysis. The plant enzymes are certainly very slow as compared to other enzymes. Plant enzymes are form I, and assemble as L8S8 complexes, whereas form II and form III enzymes, found in dinoflagellates, bacteria and archea, are complexes of the large subunit only. Here, Davidi et al. queried thousands of genomic sequences to identify about 33,000 diverse rubiscos from across the domains of life and identified more than one hundred that they expressed in E. coli for catalytic assays. They found several that are significantly faster than the plant enzymes. The fastest enzyme they characterized is from a bacterium Gallionella sp. that lives in a very low oxygen environment. (Summary by Mary Williams @PlantTeaching) EMBO J. 10.15252/embj.2019104081
This is a fascinating and very well-written paper that investigates the diversity of rubisco’s kinetic properties. Rubisco’s relationship with its substrate CO2 is complicated by its relationship with O2, and it has often been suggested that for this reason rubisco is locked into a slow rate of catalysis. The plant enzymes are certainly very slow as compared to other enzymes. Plant enzymes are form I, and assemble as L8S8 complexes, whereas form II and form III enzymes, found in dinoflagellates, bacteria and archea, are complexes of the large subunit only. Here, Davidi et al. queried thousands of genomic sequences to identify about 33,000 diverse rubiscos from across the domains of life and identified more than one hundred that they expressed in E. coli for catalytic assays. They found several that are significantly faster than the plant enzymes. The fastest enzyme they characterized is from a bacterium Gallionella sp. that lives in a very low oxygen environment. (Summary by Mary Williams @PlantTeaching) EMBO J. 10.15252/embj.2019104081