Glyphosate Resistance Decoded: the reference sequence of the extrachromosomal DNA Replicon in Amaranth
Decades of research in evolutionary genetics has shown that genomic plasticity, in particular variation in gene copy number, is a favored mechanism to provide rapid adaptation to adverse environmental conditions through increase in gene dosage. Gene amplification has been observed across kingdoms and often is supported by the accumulation of extrachromosomal circular DNA (eccDNA), ranging in size from a few hundred base pairs to hundreds of kilobases.
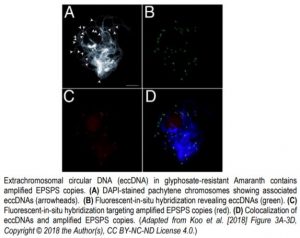
Glyphosate is a broadly used herbicide for invasive weed control in domestic and agricultural contexts. Its activity relies on the inhibition of a critical gene involved in amino acid synthesis, 5-enol pyruvylshikimate-3-phosphate synthase (EPSPS). Intensive use of the herbicide has resulted in the rapid emergence of glyphosate-resistant weed species that may reduce yields and increase weed-control costs in current agricultural settings. The molecular basis of glyphosate resistance in Palmer amaranth (Amaranthus palmeri) relies on an increase in EPSPS copy number that counteracts shikimate accumulation and loss of aromatic amino acids upon glyphosate treatment. The EPSPS amplification is intimately linked to the activity of a self-replicating, heritable eccDNA replicon (Koo et al., 2018) (Figure). The eccDNA replicon harbors several other genes, transposable elements, and arrays of highly-structured sequences that may contribute to the molecular behavior of the unit. Such complexity has so far hindered a full resolution of the eccDNA replicon.
 In a new study, Molin et al. (2020) fill this gap by providing a single-molecule-based reference sequence for the eccDNA replicon. The circular assembly of the unit reaches 399,435 bp and illuminates the features facilitating the emergence, maintenance, and transmission of the eccDNA Replicon. The reference sequence identifies 59 genes including EPSPS, 41 of which show transcriptional up-regulation upon glyphosate treatment of resistant plants. Among the most expressed genes after glyphosate exposure are EPSPS, together with genes coding for proteins harboring Plant Mobile Domains (PMD) and genes encoding for proteins with Zinc-Finger (ZF) domains. PMD proteins are thought to be derived from “domesticated” transposable elements and are involved in genome stability, possibly the stability of the eccDNA replicon itself, while ZF proteins mediate DNA-DNA interactions and may facilitate the chromosomal tethering of the unit. The single-molecule sequencing approach further facilitated the resolution of the complex arrays of repetitive sequences distributed along the replicon, and identified a high density of transposons in the immediate vicinity of EPSPS with potential implications on the genesis and behavior of the eccDNA replicon. To shine a light on the origins of the eccDNA replicon, the authors aligned the new A.palmeri replicon reference sequence to the genomes of the closely-related species waterhemp (Amaranthus tuberculatus) and grain amaranth (Amaranthus hypocondriacus). The limited levels of synteny and collinearity identified between genomes for the genes contained within the eccDNA replicon argue in favour of intra-genomic recombination being at the basis of the replicon formation.
In a new study, Molin et al. (2020) fill this gap by providing a single-molecule-based reference sequence for the eccDNA replicon. The circular assembly of the unit reaches 399,435 bp and illuminates the features facilitating the emergence, maintenance, and transmission of the eccDNA Replicon. The reference sequence identifies 59 genes including EPSPS, 41 of which show transcriptional up-regulation upon glyphosate treatment of resistant plants. Among the most expressed genes after glyphosate exposure are EPSPS, together with genes coding for proteins harboring Plant Mobile Domains (PMD) and genes encoding for proteins with Zinc-Finger (ZF) domains. PMD proteins are thought to be derived from “domesticated” transposable elements and are involved in genome stability, possibly the stability of the eccDNA replicon itself, while ZF proteins mediate DNA-DNA interactions and may facilitate the chromosomal tethering of the unit. The single-molecule sequencing approach further facilitated the resolution of the complex arrays of repetitive sequences distributed along the replicon, and identified a high density of transposons in the immediate vicinity of EPSPS with potential implications on the genesis and behavior of the eccDNA replicon. To shine a light on the origins of the eccDNA replicon, the authors aligned the new A.palmeri replicon reference sequence to the genomes of the closely-related species waterhemp (Amaranthus tuberculatus) and grain amaranth (Amaranthus hypocondriacus). The limited levels of synteny and collinearity identified between genomes for the genes contained within the eccDNA replicon argue in favour of intra-genomic recombination being at the basis of the replicon formation.
Manipulation of EPSPS dosage through copy number variation appears to be a favored target for the rapid emergence of glyphosate resistance in weeds. Such a strong permissivity to structural variation has been observed for other fitness-related genes such as disease-resistance genes in pepper (Kim et al., 2017) or the flowering regulator FLC in Arabidopsis thaliana (Quadrana et al., 2019). The rapid emergence and diversification of these highly-mutable loci might be the key for adaptive evolution in plants.
Matthias Benoit
Howard Hughes Medical Institute
Cold Spring Harbor Laboratory
ORCID: 0000-0002-3958-3173
REFERENCES
Kim, S. et al. (2017). New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 18: 1–11.
Koo, D.H., Molin, W.T., Saski, C.A., Jiang, J., Putta, K., Jugulam, M., Friebe, B., and Gill, B.S. (2018). Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri. Proc. Natl. Acad. Sci. U. S. A. 115: 3332–3337.
Quadrana, L., Etcheverry, M., Gilly, A., Caillieux, E., Madoui, M.A., Guy, J., Bortolini Silveira, A., Engelen, S., Baillet, V., Wincker, P., Aury, J.M., and Colot, V. (2019). Transposition favors the generation of large effect mutations that may facilitate rapid adaption. Nat. Commun. 10: 1–10.



