Gibberellin signaling in plants – Entry of a new microRNA player
Raimund Nagel
University of Leipzig
[email protected]
Gibberellins (GA) are essential hormones that control plant development as major regulators of stem elongation, germination, dormancy, flowering, leaf senescence and fruit development (Hedden and Thomas, 2012). Their biosynthesis and signaling pathways are well established; however, the regulatory networks that modulate signaling remain less explored (Hedden and Sponsel, 2015). Bioactive GAs are perceived through a specific receptor (GID1), which in its GA-free state is bound to a DELLA protein, a master regulator of GA signaling. Upon GA binding, the DELLA protein is ubiquitinylated and ultimately degraded in the proteasome, releasing sequestered transcription factors that now induce transcription of GA targets. Rice (Oryza sativa) is a well-established model system for GA research that has the advantage of having only one DELLA (SLR1) in contrast to the five DELLA proteins in Arabidopsis (Van De Velde et al., 2017). Therefore, rice is an ideal system in which to study GA signaling.
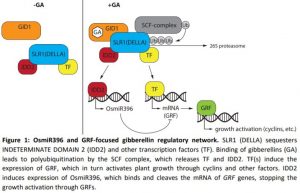 In this issue of Plant Physiology, Lu et al. (2020) set out to investigate how GA signaling is modulated by the microRNA OsmiR396m. OsmiR396 is also well conserved in other plants and has roles in several different pathways (Liebsch and Palatnik, 2020). OsmiRNA396 was previously shown to interact with GA-responsive growth regulating factors (GRF). Rice GRFs are regulated by OsmiR396 through cleavage of the mRNA at the complementary site. When OsmiR396 was overexpressed, the resulting plants had a dwarf phenotype, caused by fewer and to a lesser degree smaller individual cells. Surprisingly the dwarf phenotype could not be rescued by GA application. Expression studies showed that wild-type plants upregulate GRF1 after GA application and reduce expression of OsmiRNA396, while OsmiR396-overexpressing plants do not increase expression of GRF1 after GA application. Overexpression of OsmiR396 also decreased expression of cell-cycle-related genes, like those encoding cyclins and cyclin-dependent-kinase, explaining the lower cell numbers.
In this issue of Plant Physiology, Lu et al. (2020) set out to investigate how GA signaling is modulated by the microRNA OsmiR396m. OsmiR396 is also well conserved in other plants and has roles in several different pathways (Liebsch and Palatnik, 2020). OsmiRNA396 was previously shown to interact with GA-responsive growth regulating factors (GRF). Rice GRFs are regulated by OsmiR396 through cleavage of the mRNA at the complementary site. When OsmiR396 was overexpressed, the resulting plants had a dwarf phenotype, caused by fewer and to a lesser degree smaller individual cells. Surprisingly the dwarf phenotype could not be rescued by GA application. Expression studies showed that wild-type plants upregulate GRF1 after GA application and reduce expression of OsmiRNA396, while OsmiR396-overexpressing plants do not increase expression of GRF1 after GA application. Overexpression of OsmiR396 also decreased expression of cell-cycle-related genes, like those encoding cyclins and cyclin-dependent-kinase, explaining the lower cell numbers.
In a large yeast one-hybrid screen, the promotor of OsmiR396 was tested against a rice cDNA library. All positive clones were identified as O. sativa INDETERMINATE DOMAIN 2 (OsIDD2), a four-zinc finger motif containing protein, with binding to the promotor confirmed through electrophoretic mobility shift assays. Plants overexpressing OsIDD2 showed a dwarf phenotype with higher expression of OsmiR396 and lower expression of GRF1. GA application reversed these expression patterns and rescued the dwarf phenotype. RNAi knock-down plants showed the opposite phenotype and expression pattern, but application of an inhibitor of GA biosynthesis did not lead to dwarf plants, leaving open the question of how OsIDD2 is regulated. It was hypothesized that SLR1 and OsIDD2 interact on the protein level, which was demonstrated thoroughly through three different experiments: yeast two-hybrid, GST pulldown assays, and co-immunoprecipitation.
These results suggest a negative feedback loop in GA signaling that starts with GA binding to the receptor. Subsequent DELLA (SLR1) degradation releases OsIDD2, which in turn binds to the promotor of OsmiR396 and increases its expression. OsmiR396 itself then cleaves the mRNA of GA responsive GRF genes, thereby stopping the GA signaling cascade and growth promotion (Figure 1). The question remains of how the timing of GRF and OsmiR396 expression and activity are regulated.
This study emphasizes the complexity of GA signaling, with numerous negative regulators that must be countered. At the same time, this complex regulation might allow us to intentionally manipulate GA effects in different tissues and at very specific time points to maximize production of edible plant parts without hampering the plant defense reaction. Evidence for an apparent antagonism between growth and defense can especially be seen in rice, as two economically important pathogens independently evolved gibberellin biosynthetic pathways and produce gibberellins upon infection. One is the fungus Gibberella fujikuroi, from which gibberellins were originally isolated, and the other is the bacteria Xanthomonas oryzae pv. orizicola (Tudzynski et al., 2003; Nagel et al., 2017). For these reasons successful engineering of GA signaling will be a highly complex task, but could also be very rewarding.
REFERENCES
Hedden P, Sponsel V (2015) A Century of Gibberellin Research. Journal of Plant Growth Regulation 34: 740-760
Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444: 11-25
Liebsch D, Palatnik JF (2020) MicroRNA miR396, GRF transcription factors and GIF co-regulators: a conserved plant growth regulatory module with potential for breeding and biotechnology. Current Opinion in Plant Biology 53: 31-42
Lu Y, Feng Z, Meng Y, Bian L, Xie H, Mysore KS, Liang J-S (2020) SLENDER RICE1 and Oryza sativa INDETERMINATE DOMAIN2 Regulating OsmiR396 Is Involved in Stem Elongation. Plant Physiology: pp.01008.02019 https://doi.org/10.1104/pp.19.01008
Nagel R, Turrini PC, Nett RS, Leach JE, Verdier V, Van Sluys MA, Peters RJ (2017) An operon for production of bioactive gibberellin A4 phytohormone with wide distribution in the bacterial rice leaf streak pathogen Xanthomonas oryzae pv. oryzicola. New Phytologist 214: 1260-1266
Tudzynski B, Mihlan M, Rojas MC, Linnemannstons P, Gaskin P, Hedden P (2003) Characterization of the final two genes of the gibberellin biosynthesis gene cluster of Gibberella fujikuroi: des and P450-3 encode GA4 desaturase and the 13-hydroxylase, respectively. J Biol Chem 278: 28635-28643
Van De Velde K, Ruelens P, Geuten K, Rohde A, Van Der Straeten D (2017) Exploiting DELLA Signaling in Cereals. Trends Plant Sci 22: 880-893



