Genome editing to enhance broad-spectrum disease resistance in rice without compromising yield
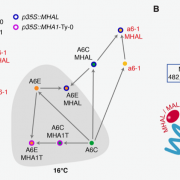
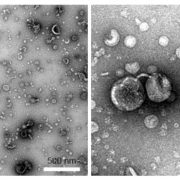
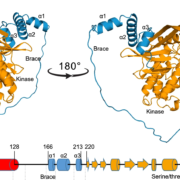
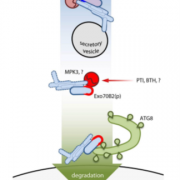
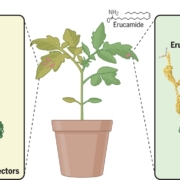
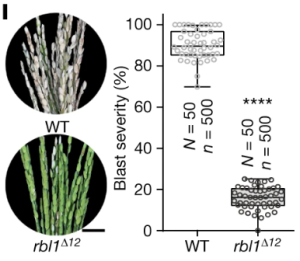 Developing broad-spectrum disease resistant crops without compromising the yield has been a persistent challenge in maintaining global food security. In a breakthrough study, Sha et al. developed a promising multi-pathogen resistant rice mutant that retains the crop’s yield, using forward genetic screen and genome editing approaches. The research journey began with the identification of a group of lesion mimic mutants (LMM) from a mutagenized rice population. Among these, a mutant named resistance to blast1 (rbl1) displayed resistance to both fungal and bacterial pathogens but had the drawback of low fertility and yield. The causal mutation was mapped to be a 29-bp deletion in the RBL1 gene, encoding for the CDP-DAG synthase (CDS). CDP-DAG is a precursor for phosphatidylinositol (PI) and its downstream products PI3P, PI4P, and PI(4,5)P2. The rbl1 mutation led to a decrease of PI(4,5)P2, a key player in fungal-plant interactions. To engineer a rbl1 allele without yield loss, Sha et al. strategically applied genome editing to target different sites of RBL1. They successfully identified a new allele, rbl1△12, carrying a 12-bp deletion. Impressively, rbl1△12 not only demonstrated resistance to a range of fungal and bacterial pathogens, but also maintained wild-type yield in small-scale field trial. This study represents a significant step forward in the quest for durable disease-resistant crops. (Summary by Xiaohui Li @Xiao_hui_Li) Nature 10.1038/s41586-023-06205-2
Developing broad-spectrum disease resistant crops without compromising the yield has been a persistent challenge in maintaining global food security. In a breakthrough study, Sha et al. developed a promising multi-pathogen resistant rice mutant that retains the crop’s yield, using forward genetic screen and genome editing approaches. The research journey began with the identification of a group of lesion mimic mutants (LMM) from a mutagenized rice population. Among these, a mutant named resistance to blast1 (rbl1) displayed resistance to both fungal and bacterial pathogens but had the drawback of low fertility and yield. The causal mutation was mapped to be a 29-bp deletion in the RBL1 gene, encoding for the CDP-DAG synthase (CDS). CDP-DAG is a precursor for phosphatidylinositol (PI) and its downstream products PI3P, PI4P, and PI(4,5)P2. The rbl1 mutation led to a decrease of PI(4,5)P2, a key player in fungal-plant interactions. To engineer a rbl1 allele without yield loss, Sha et al. strategically applied genome editing to target different sites of RBL1. They successfully identified a new allele, rbl1△12, carrying a 12-bp deletion. Impressively, rbl1△12 not only demonstrated resistance to a range of fungal and bacterial pathogens, but also maintained wild-type yield in small-scale field trial. This study represents a significant step forward in the quest for durable disease-resistant crops. (Summary by Xiaohui Li @Xiao_hui_Li) Nature 10.1038/s41586-023-06205-2