Follow that Protein: SNAP-tagging Permits High-resolution Protein Localization
Protein analysis relies heavily on the production of ‘tagged’ proteins, i.e. recombinant proteins that contain both a functional peptide sequence and a peptide or chemical label (the ‘tag’). Common tags include fluorescent proteins (FPs), which make tagged proteins easier to detect, as well as tags that bind preferentially to specific substrates, which facilitates protein purification. However, new chemical dye tags have been developed that offer notable advantages over fluorescent protein tags, including smaller size, higher quantum yield and photostability, more specific timing of the initiation of fluorescence, and a greater variety of dyes that can be used. One of these new methods is SNAP-tagging, a method of protein labelling that involves covalently bonding a target protein to a ‘SNAP-tag’, a modified version of human O6-alkylguanine-DNA-alkyltransferase that can take a variety of dyes and fluorophores as substrates (Juillerat et al., 2003, Keppler et al., 2003). Despite the fact that SNAP-tagging has been used extensively to study endocytosis in human and animal systems (Thorn, 2017), this technique is not commonly used in studies of plant proteins, possibly because the synthetic dyes involved in SNAP-tagging may not effectively pass through plant cell walls.
 In their recent paper, Iwatate et al. (2020) attempted to establish that SNAP-tagging can be effectively used to study plant proteins in vivo. To demonstrate the usefulness of SNAP-tagging in plant systems, the authors performed five key tests. First, they determined whether plant cells were permeable to dyes of interest. This was assessed by examining the permeability of tobacco BY-2 cells to an array of 32 different fluorescent dyes. The authors identified 23 dyes that were able to pass through the cell wall and enter the cell within one minute at pH 5.8. Eight dyes did not enter the cytosol at all, and three dyes—Rhodamine Green, Rhodamine 123, and PREX710—accumulated only to a very slight degree. Three dyes used for SNAP-tagging in animal systems—Surface Alexa Fluor 488, SNAP-Cell TMR-Stars, and SNAP-Cell-647-SiR—were found to be able to permeate BY-2 cells either in their native forms or in benzylguanine-conjugated forms.
In their recent paper, Iwatate et al. (2020) attempted to establish that SNAP-tagging can be effectively used to study plant proteins in vivo. To demonstrate the usefulness of SNAP-tagging in plant systems, the authors performed five key tests. First, they determined whether plant cells were permeable to dyes of interest. This was assessed by examining the permeability of tobacco BY-2 cells to an array of 32 different fluorescent dyes. The authors identified 23 dyes that were able to pass through the cell wall and enter the cell within one minute at pH 5.8. Eight dyes did not enter the cytosol at all, and three dyes—Rhodamine Green, Rhodamine 123, and PREX710—accumulated only to a very slight degree. Three dyes used for SNAP-tagging in animal systems—Surface Alexa Fluor 488, SNAP-Cell TMR-Stars, and SNAP-Cell-647-SiR—were found to be able to permeate BY-2 cells either in their native forms or in benzylguanine-conjugated forms.
Second, the authors tested the self-labelling potential of these three SNAP-tagging dyes by transforming BY-2 cells with a construct encoding a SNAP-tagged α-tubulin 5 protein (pUBQ10:SNAP-TUA5), and determining whether cytoskeletal structures containing α-tubulin could be visualized after incubation with any of the three dyes. They observed specific, efficient labelling of microtubules for each dye, indicating that SNAP-tagging with these dyes is feasible. The authors also captured microtubule transitions in a short film, included in the Supplemental Material, which is visually captivating and may be of interest to readers.
The third set of experiments repeated the α-tubulin labelling experiment in Arabidopsis thaliana. Using the same SNAP-TUA5 construct, they were again able to visualize cytoskeletal structures, including spindles and phragmoplasts, in A. thaliana seedlings. They also examined whether SNAP-tagged proteins posed a toxicity risk by tracking the mitotic cell division in etiolated hypocotyl cells infiltrated with SNAP-tagged tubulin and those infiltrated with tubulin tagged with an FP, mCherry-TUA5. They found no significant difference in the time from nuclear envelope breakdown to phragmoplast initiation, suggesting that SNAP-tagged proteins do not contribute to cellular toxicity more than comparable FPs.
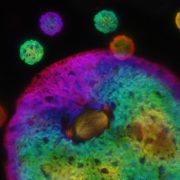
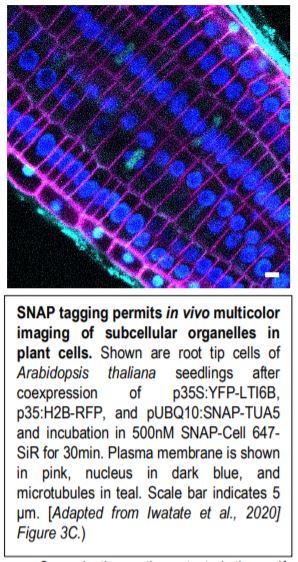
Fourth, the authors demonstrated one of the major advantages of SNAP-tagging as a method of protein visualization: the ability to use the specific spectral properties of different chemical dyes to permit in vivo multicolor imaging of subcellular structures (see Figure). Finally, the authors attempted to perform SNAP-tagging of de novo PIN2, a specific auxin transporter regulating gravitropism, and to track where these proteins moved within the cell. By determining the fluorescence ratio of SNAP-tagged proteins to an FP-tagged cell membrane-impermeable probe (DRBG488), they were able to demonstrate that the SNAP-tagged protein permitted the effective monitoring of endocytosis, intracellular trafficking, and the delivery of newly synthesized protein to the cell plate, and permitted the direct visualization (via double tagging) of multiple pools of the same protein within a single cell.
Taken together, these data suggest that the in vivo targeted labelling of proteins with small molecule chemical dyes is possible in plant systems as it is in animal systems. Here the authors demonstrate that SNAP-tagging can be effectively be used for a variety of novel experiments, including multicolor labelling and the observation of cytoskeletal dynamics. This is a valuable achievement because chemical dye labelling offers more precise qualification and subcellular localization relative to covalent FP-based tagging.
William Hughes
Department of Ecology, Environment, and Plant Sciences,
Stockholm University
Svante Arrhenius väg 20
114 18 Stockholm
ORCID: 0000-0003-4142-5279
REFERENCES
Iwatate R, Yoshinari A, Yagi N, Grzybowski M, Ogasawara H, Kamiya M, Komatsu T, Taki M, Yamaguschi S, Frommer W, and Nakamura M. (2020) Covalent Self-labeling of Tagged Proteins with Chemical Fluorescent Dyes in BY-2 Cells and Arabidopsis Seedlings. The Plant Cell. In press. DOI: https://doi.org/10.1105/tpc.20.00439
Juillerat A, Gronemeyer T, Keppler A, Gendrezig S, Pick H, Vogel H, and Johnsson K. 2003. Directed evolution of O6-alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo. Chemical Biology 10(4):313-317. DOI: 10.1016/S1074-5521(03)00068-1
Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, and Johnsson K. (2003). A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nature Biotechnology 21: 86–89. DOI: 10.1038/nbt765
Thorn K. (2017). Genetically encoded fluorescent tags. Molecular Biology of the Cell, 28:848–857. DOI: 10.1091/mbc.E16-07-0504