Evolution of tetraploid meiosis (PNAS)
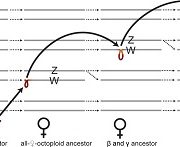
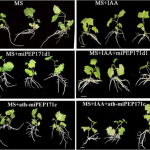
 Genome duplications are common in plants and thought to be an important contributor to evolutionary innovations, but the increase in ploidy that results from a genome duplication also presents challenges for reproduction. Because there are four sets of homologous chromosomes in the derived tetraploid (instead of two in the diploid ancestor), establishing one-to-one pairing between homologs in meiosis can be a problem. Failure to form such “bivalents” leads to segregation errors and reduced fertility. This would prevent newly formed tetraploids (neopolyploids) from becoming established. However, the plant world is full of polyploid species, meaning that evolution in many cases is able to overcome this obstacle. One way to understand how this could happen is to compare the meiotic division machinery between diploids and tetraploids. Arabidopsis arenosa is an ideal system for this type of investigation because there are both diploid (ancestral) and tetraploid (derived) subpopulations available for comparison. Two meiosis genes, ASY1 and ASY3 are under selection in the tetraploid, suggesting that they may play important roles. These two genes both code for proteins that localize to the so-called chromosome axis, which is central to meiotic pairing. In this paper, Morgan et al. introduce the ASY1 and ASY3 alleles from the diploid ancestor into the tetraploid and notice how this changes several aspects of chromosome behavior during meiotic pairing. In particular, they observe an increased frequency of harmful multivalent pairings. However, the overall effect size was modest. The authors predict that small changes in many genes (including ASY1/3) are needed to cope with the challenges of tetraploid meiosis. (Summary by Frej Tulin @FrejTulin) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1919459117
Genome duplications are common in plants and thought to be an important contributor to evolutionary innovations, but the increase in ploidy that results from a genome duplication also presents challenges for reproduction. Because there are four sets of homologous chromosomes in the derived tetraploid (instead of two in the diploid ancestor), establishing one-to-one pairing between homologs in meiosis can be a problem. Failure to form such “bivalents” leads to segregation errors and reduced fertility. This would prevent newly formed tetraploids (neopolyploids) from becoming established. However, the plant world is full of polyploid species, meaning that evolution in many cases is able to overcome this obstacle. One way to understand how this could happen is to compare the meiotic division machinery between diploids and tetraploids. Arabidopsis arenosa is an ideal system for this type of investigation because there are both diploid (ancestral) and tetraploid (derived) subpopulations available for comparison. Two meiosis genes, ASY1 and ASY3 are under selection in the tetraploid, suggesting that they may play important roles. These two genes both code for proteins that localize to the so-called chromosome axis, which is central to meiotic pairing. In this paper, Morgan et al. introduce the ASY1 and ASY3 alleles from the diploid ancestor into the tetraploid and notice how this changes several aspects of chromosome behavior during meiotic pairing. In particular, they observe an increased frequency of harmful multivalent pairings. However, the overall effect size was modest. The authors predict that small changes in many genes (including ASY1/3) are needed to cope with the challenges of tetraploid meiosis. (Summary by Frej Tulin @FrejTulin) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1919459117