Down the Rabbit Hole: The Hidden World of Gene Regulation Within the Nucleus
Lee and Bailey-Serres uncover multiple aspects of nuclear and whole-cell epigenetic and post-transcriptional gene regulation in Arabidopsis. Plant Cell https://doi.org/10.1105/tpc.19.00463
By T. A. Lee and J. Bailey-Serres
Background: Plant cells require oxygen to generate ATP, similar to our own living cells. Cells of dense tissues or flooded organs can experience hypoxia. This situation causes a low-energy crisis that necessitates inefficient use of sugars to produce ATP. How does a plant cell minimize energy usage and a loss of resources? How does it rapidly recover once oxygen becomes available again? We showed previously that plants manage energy during this stress by minimizing translation, using only the most highly upregulated gene transcripts for protein synthesis. We knew that transcriptional activation under hypoxia involves a group of transcription factors that are continuously synthesized but are typically only stable when oxygen levels are extremely low. However, knowledge of the dynamics of gene transcription within the nucleus, and the role of epigenetic regulation, was lacking.
Question: Gene regulation occurs in two compartments and is fine-tuned at multiple points of control. We wanted to know if there is coordination from chromatin (the epigenome) transcript synthesis, through mRNA translation, to prioritize the upregulation of critical genes.
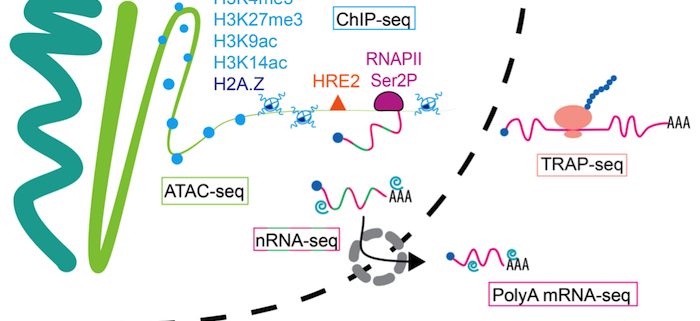
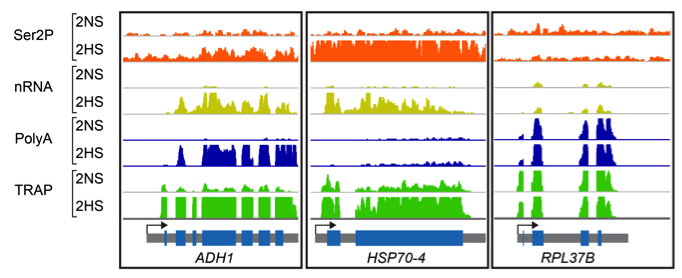 Findings: Hypoxia promotes underappreciated alterations of gene regulation within the nucleus of Arabidopsis thaliana seedlings. To unravel the complexities, we identified groups of similarly regulated genes. Genes highly upregulated at the RNA level (nuclear, polyadenylated and translated mRNA), termed coordinate Hypoxia Regulated Genes (co-HRGs), had histone and RNAPII signatures that said “transcribe and translate now!”. Genes associated with heat and general acute stress had a different signature. These said transcribe now and translate later, following prolonged hypoxia or upon re-oxygenation. The nuclear regulation of genes associated with growth, such as those encoding the proteins of ribosomes, were interesting as well. RNAPII remained engaged on these genes, producing transcripts that appear to remain in the nucleus until reoxygenation. A JBrowse session containing the data can be found at: https://www.baileyserreslab.org/data.
Findings: Hypoxia promotes underappreciated alterations of gene regulation within the nucleus of Arabidopsis thaliana seedlings. To unravel the complexities, we identified groups of similarly regulated genes. Genes highly upregulated at the RNA level (nuclear, polyadenylated and translated mRNA), termed coordinate Hypoxia Regulated Genes (co-HRGs), had histone and RNAPII signatures that said “transcribe and translate now!”. Genes associated with heat and general acute stress had a different signature. These said transcribe now and translate later, following prolonged hypoxia or upon re-oxygenation. The nuclear regulation of genes associated with growth, such as those encoding the proteins of ribosomes, were interesting as well. RNAPII remained engaged on these genes, producing transcripts that appear to remain in the nucleus until reoxygenation. A JBrowse session containing the data can be found at: https://www.baileyserreslab.org/data.
Next steps: This study raises new questions about stress-activated and dampened transcription. Are transcription factors poised, RNAPII paused, or fully processed mRNAs retained in the nucleus during an acute stress? What are the roles of chromatin-modifying enzymes? Further study may identify regulatory processes important for stress resilience with use in synthetic biology.
Travis A. Lee and Julia Bailey-Serres (2019). Integrative Analysis from the Epigenome to Translatome Uncovers Patterns of Dominant Nuclear Regulation during Transient Stress. Plant Cell. https://doi.org/10.1105/tpc.19.00463.
Key words: Hypoxia, epigenetics, post-transcriptional gene regulation, stress-recovery