CsCRC regulates cucumber fruit length
Che, Pan, Liu, Li et al. provide insight into fruit length variation in cucumber.
By Gen Chea,e, Yupeng Panb, Xiaolan Zhanga,*
aState Key Laboratories of Agrobiotechnology, Beijing Key Laboratory of Growth and Developmental Regulation for Protected Vegetable Crops, MOE Joint Laboratory for International Cooperation in Crop Molecular Breeding, China Agricultural University, Beijing, 100193, China;
bHorticulture Department, University of Wisconsin-Madison, 1575 Linden Drive, Madison, WI 53706, USA;
cSchool of Life Science, Key Laboratory of Herbage & Endemic Crop Biology, Ministry of Education, Inner Mongolia University, Hohhot 010070, China
Background: Cucumber is an important vegetable crop cultivated worldwide. The fruit length in cucumber varies from 5 to 60 cm in different germplasms, and thus serves as a key domestication trait affecting yield and appearance quality. Although several regulators and multiple QTLs controlling fruit length have been identified, the natural variations and molecular mechanisms underlying fruit length divergence are largely unknown.
Question: How can cucumber vary so widely in its fruit length? FS5.2 was previously identified as a major-effect QTL specifying fruit length in cucumber, while the underlying controlling gene and its regulating mechanism are unclear.
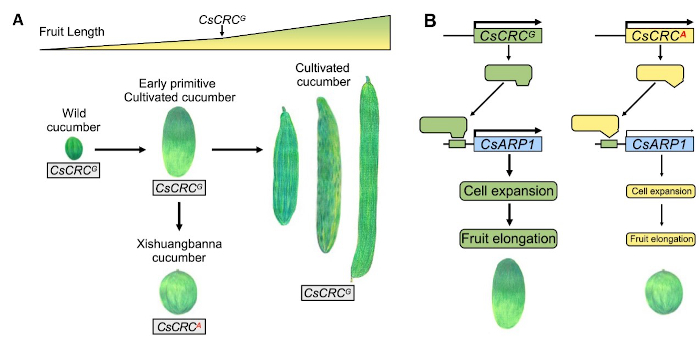
Findings: A nonsynonymous SNP (G to A) in CsCRC was identified to control the fruit length variation caused by the FS5.2 locus. Allelic analysis of 165 cucumber accessions showed that the CsCRCA allele only exists in 7 Xishuangbanna cucumbers with short or round fruits, while the CsCRCG allele was shared by all the remaining wild and cultivated cucumbers. Replacing the CsCRCG allele with CsCRCA by backcrossing resulted in a 34~39% reduction in fruit length. Genetic transformation-based functional characterization revealed that CsCRCG is a positive regulator of fruit length elongation and cell size expansion. CsCRCG, but not CsCRCA, targets the downstream auxin-responsive protein gene CsARP1 (a positive mediator of fruit length), to promote its expression and stimulate fruit elongation in cucumber.
Next step: Manipulation of fruit length can be achieved by either modulating the expression levels of CsCRCG or utilizing different CsCRC alleles (CsCRCG or CsCRCA) in cucumber breeding practices. The regulatory mechanism of CsARP1 and its relation with auxin await further characterization in plants.
Gen Che, Yupeng Pan, Xiaofeng Liu, Min Li, Jianyu Zhao, ShuangshuangYan, Yuting He, Zhongyi Wang, Zhihua Cheng, Weiyuan Song, Zhaoyang Zhou, Tao Wu , Yiqun Weng, Xiaolan Zhang. (2023). Natural variation in CRABS CLAW contributes to fruit length divergence in cucumber https://doi.org/10.1093/plcell/koac335