Corn ChIPs and RNA-seq: Researchers Dip into Advanced Tools and Resources to Examine bZIP Transcription Factor Function in the Maize Endosperm
The endosperm of maize (Zea mays) seeds undergoes a complicated developmental program that ends with the production of massive amounts of storage compounds, particularly carbohydrates, but also including zein storage proteins (reviewed in Li and Berger, 2012; Hannah and Boehlein, 2017; Larkins et al., 2017). In two recent papers from The Plant Cell, the authors take advantage of the sets of advanced tools and resources now available in maize (Zea mays) to probe the functions of bZIP transcription factors in development and function of the endosperm.
Maize grains do not have adequate lysine and tryptophan for use as food and feed and 2 the major storage protein 7-kD γ-zein lacks these amino acids entirely. Downregulation of γ-zein, and compensation by other storage proteins, improves the amino acid balance of maize grains. Li et al. (2018) examine the regulation of the gene encoding 27-kD γ-zein, which is known to be regulated by multiple transcription factors, including Opaque-2 (O2). However, the full details of this regulation, and how all these transcription factors feed into the final transcriptional output remain unclear. To address this, the authors use electrophoretic mobility shift assays to find factors that bind to the 27-kD γ-zein promoter region. They use nuclear proteins from developing maize endosperm and look for shifts with labeled promoter fragments. In addition to known factors such as O2, they found a shift caused by an unknown protein. They identified that protein as ZmbZIP22 by DNA-affinity pull-down (with the shifted fragment), gel separation and silver staining, followed by liquid chromatography-mass spectrometry.
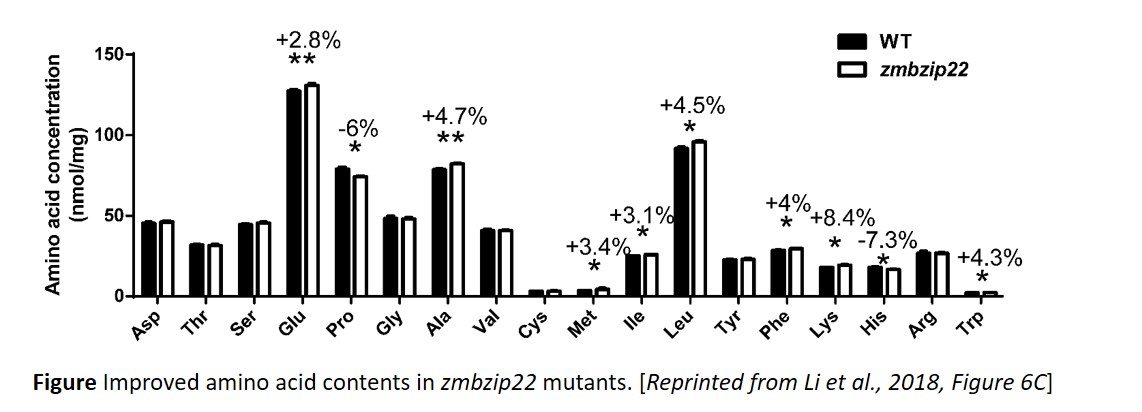
ZmbZIP22 directly activated expression from the 27-kD γ-zein promoter and zmbzip22 mutants had reduced 27-kD γ-zein levels. Despite the reduction in γ-zein, total protein remained the same in zmbzip22 mutants as in wild type and the change in protein distribution resulted in an improvement in amino acid contents (figure). By a combination of RNA sequencing in zmbzip22 mutants and chromatin immunoprecipitation sequencing (ChIP-seq) to identify ZmbZIP22 binding sites, the authors defined the sets of genes directly or indirectly activated or repressed by ZmbZIP22. Finally, since bZIP transcription factors function as homodimers or heterodimers, the authors examine the physical and functional interactions between ZmbZIP22 and the other transcription factors that regulate zein expression.
In another innovative study of bZIP transcription factors in maize endosperm, Zhan et al. (2018) examine the regulatory network of O2, which (as mentioned above) includes the genes encoding 27-kD γ-zein and a host of other endosperm-expressed proteins. One outstanding question is which of these genes are regulated directly and which are regulated indirectly by O2. To address this, the authors used RNA sequencing to identify genes differentially expressed in the endosperm between wild type and o2 mutants. Comparison of the 1,863 differentially expressed genes and the 3,282 genes with nearby O2 binding sites determined by ChIP-seq identified a surprisingly low number of direct O2 targets. Indeed, only 186 genes had nearby O2 binding sites and were differentially expressed. These direct targets included most of the known targets of O2 (confirming the soundness of the approach), as well as myriad genes involved in the accumulation of storage carbohydrates and lipids, and most of the zein genes. Moreover, the authors compared the temporal expression patterns of the direct O2 targets with the expression of O2 itself during endosperm development in wild-type and mutant seeds. This revealed different modes of gene activation by O2 and a role for other transcription factors in maintaining the maximal expression of the target genes during development. The direct targets of O2 included 15 transcription factors, of which 5 are bZIP proteins, suggesting these O2-regulated bZIP TFs might be additional regulators of maize endosperm development. Finally, previous studies had identified several binding motifs for O2. However, only about 20% of the O2 binding sites identified by ChIP-seq contained these motifs, indicating the existence of other O2 binding motifs. Indeed, analysis of the O2 ChIP data identified additional motifs that are likely O2 binding sites.
Advanced tools and resources for gene expression studies are now available in maize and studies using these tools have provided important insight into the complex regulatory networks acting in the endosperm. Moreover, the application of multiple tools to a given problem such as gene regulation by O2 has shown that one tool (i.e. transcriptome assays) does not provide a complete picture of the complexity of gene regulation—only application of several tools, such as transcriptome and binding site analysis, gives a clearer picture. Indeed, the emerging availability of resources and tools for maize (and many other systems beyond Arabidopsis) has enabled innovative research such as that presented by these two studies. Furthermore, these studies have implications for improving human nutrition, although additional studies will be required to identify the best combination of alleles for optimal grain quality without adverse agronomic effects.
REFERENCES
Hannah, L.C., and Boehlein, S. (2017). 12 Starch Biosynthesis in Maize Endosperm. Maize Kernel Development, 149.
Larkins, B.A., Wu, Y., Song, R., and Messing, J. (2017). 14 Maize Seed Storage Proteins. Maize Kernel Development, 175.
Li, C., Yue, Y., Chen, H., Qi, W., and Song, R. (2018) The ZmbZIP22 Transcription Factor Regulates 27-kD γ-Zein Gene Transcription during Maize Endosperm Development. Plant Cell 10.1105/tpc.18.00422
Li, J., and Berger, F. (2012) Endosperm: food for humankind and fodder for scientific discoveries. New Phytologist 195, 290-305.
Zhan, J., Li, G., Ryu, C.-H., Ma, C., Zhang, S., Lloyd, A., Hunter, B.G., Larkins, B.A., Drews, G.N., Wang, X., and Yadegari, R. (2018) Opaque-2 Regulates a Complex Gene Network Associated with Cell Differentiation and Storage Functions of Maize Endosperm. Plant Cell 10.1105/tpc.18.00392.