Copy Number Variations in the Arabidopsis Genome
Żmieńko et al. generate a catalog of large copy number variations in Arabidopsis, shedding light on the genetic basis of phenotypic variation. Plant Cell https://doi.org/10.1105/tpc.19.00640
By Agnieszka Żmieńko and Marek Figlerowicz, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznań, Poland
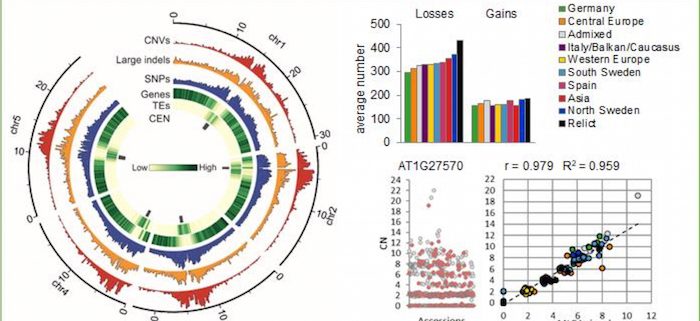
Background: The genomes of individuals of a single species are not identical. There are genomic differences of various types (e.g., presence, absence, duplication, sequence alteration, or change in the location of a DNA fragment in one genome compared to another) and sizes (they may involve any DNA fragment from 1 bp to the entire chromosome). Variations in the number of copies of large DNA fragments (typically 500 bp or longer), named CNVs, may directly affect the structure and number of the genes they overlap. This in turn may cause phenotypic variation ranging from disease to increased adaptation of an individual with a specific CNV genotype.
Question: We wanted to identify CNVs in the Arabidopsis genome and evaluate how they affect the structure and genomic distribution of genes and transposable elements. We also wanted to test whether CNVs may be useful for genetic and functional studies.
Findings: We compared the genome sequencing data collected from 1,064 Arabidopsis accessions. We identified numerous CNVs, which are typically shorter than 20 kbp but together cover over one-third of the Arabidopsis genome. CNVs are concentrated in regions that are abundant in transposable elements and poor in protein-coding genes. Nevertheless, over 18% of genes overlap with CNVs. These genes are enriched for functions related to biotic stress responses. We determined the gene copy numbers in each accession and showed that these data are useful for population analysis in Arabidopsis. We used the CNVs to analyze population structure and reveal the genetic similarity of geographically distant accessions. We also demonstrated how variation in the number of specific genes might lead to variation at the gene transcriptional level, protein level, or phenotypic level. Additionally, our observations indicate that selective forces have opposite effects on shaping variation and the relative distribution patterns of genes and transposable elements.
Next steps: The map of CNVs in the Arabidopsis genome will help researchers explore the impact of this type of genetic polymorphism on various phenotypic traits. New gene variants that are not present in the reference genome can now be identified and studied.
Agnieszka Zmienko, Malgorzata Marszalek-Zenczak, Pawel Wojciechowski, Anna Samelak-Czajka, Magdalena Luczak, Piotr Kozlowski, Wojciech M. Karlowski, Marek Figlerowicz (2020). AthCNV: A Map of DNA Copy Number Variations in the Arabidopsis thaliana Genome. Plant Cell: DOI: https://doi.org/10.1105/tpc.19.00640