Comparing genes that govern flower development in Petunia and Arabidopsis: Evolution made a mess!
Morel et al. investigate AP1/SEP/AGL6 MADS-box transcription factor functions in Petunia. Plant Cell https://doi.org/10.1105/tpc.19.00162
By Michiel Vandenbussche
Background: The ABCE model is a simple genetic model that explains how the different floral organs in the flower (sepals, petals, stamens and carpels) acquire their identity based on the combinatorial action of 4 classes of genes (A,B,C and E). These ABCE genes were identified at the DNA level and mainly belong to the gene family of MADS-box transcription factors, a class of genes that orchestrate the activity of many other genes required to make the different floral organs. Because most flowering species share a comparable ABCE gene set in their genomes, it was logically assumed that their ABCE genes would encode the same functions, although this has not been fully tested yet. Arabidopsis, a major plant genetics research model, is the species in which these genes have been characterized in the greatest detail.
 Question: We wanted to know if these ABCE genes indeed all have the same functions in other species to better understand the evolution of these master floral regulators. We addressed this question in petunia, which is separated from Arabidopsis by ~100 million years of independent evolution.
Question: We wanted to know if these ABCE genes indeed all have the same functions in other species to better understand the evolution of these master floral regulators. We addressed this question in petunia, which is separated from Arabidopsis by ~100 million years of independent evolution.
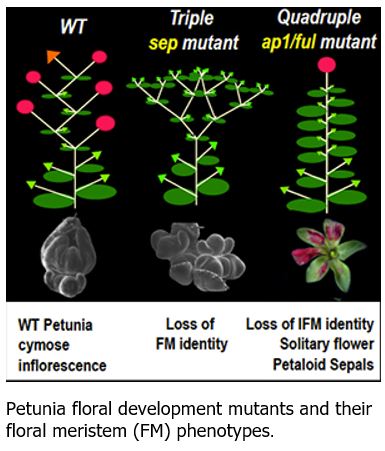
Findings: To study their function, we identified mutants for 11 petunia genes that previously were classified as A and E genes based on their sequence similarity. Our analysis shows that some of these genes indeed perform similar functions compared to their counterparts in Arabidopsis, but we also observed other cases where they clearly function differently. For example, we found that a particular class of E genes in Petunia is required to provide floral meristem identity (corresponding to the capacity of the plant’s growth tip to make flowers), while this function in Arabidopsis is associated with genes belonging to the A type. In general, our results show that the specific function of genes often cannot be accurately predicted simply by looking at their sequence. This work also shows that evolution generated greater diversity in the genetic networks driving flower development than previously thought.
Next steps: A major future challenge is to understand how such differences in gene functions may have contributed to architectural diversity between (plant) species.
Patrice Morel, Pierre Chambrier, Véronique Boltz, Sophy Chamot, Frédérique Rozier, Suzanne Rodrigues Bento, Christophe Trehin, Marie Monniaux, Jan Zethof and Michiel Vandenbussche. (2019). Divergent Functional Diversification Patterns in the SEP/AGL6/AP1 MADS-box Transcription Factor Superclade. Plant Cell. https://doi.org/10.1105/tpc.19.00162.
Key words: flower development, floral ABC model, evolution of the flower, floral diversity