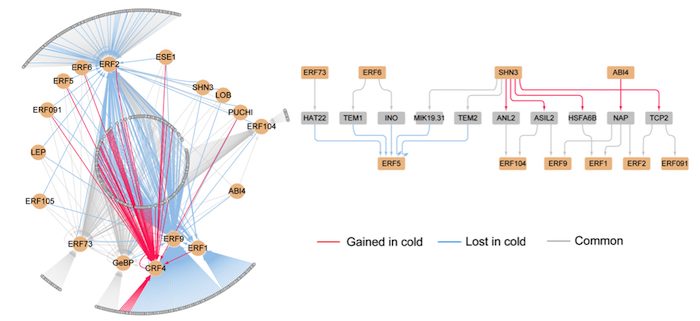
Cold-responsive Regulatory DNA Landscapes and Regulatory Networks in Grasses
Han et al. uncover regulatory DNA landscapes in root, stem, and leaf tissues of multiple grasses. Plant Cell https://doi.org/10.1105/tpc.19.00716
By Kai Wang and Jinlei Han
Fujian Agriculture and Forestry University
Background: Low temperature is a major environmental stress that affects plant growth and development and can significantly reduce crop yields under extreme conditions. Plants have evolved sophisticated regulatory systems for gene expression to help them adapt to low-temperature stress. Identifying the regulatory DNA and associated regulatory network involved in the cold-stress response is one of the most important, challenging goals of plant science and is crucial for crop improvement in the future. The DNase I-hypersensitive site (DHS) is the hallmark of active regulatory DNA and thus can be used to develop regulatory DNA maps in eukaryotes.
Question: We wanted to elucidate the regulatory DNA landscapes for different tissues of various grasses. We were curious if there was a conserved regulatory network in grasses in response to cold stress.
Findings: We developed genome-wide DHS maps for control and cold-treated leaf, stem, and root tissues of three grass species: Brachypodium distachyon, Setaria italica, and Sorghum bicolor. We found that the DHS landscapes were highly different among tissues in these species. Moreover, tissue-specific DHSs usually serve as distal control elements, which are located far away from target genes. We revealed that the DHSs were reprogrammed and showed distinct landscapes in each tissue after cold treatment, indicating that there is no conserved regulatory network among tissues within any grass. However, the DHS-derived transcription factors are largely co-located among tissues and species and include 17 motifs covering all grass taxa and all tissues examined in this study. Finally, we constructed a cold-responsive regulatory network for each tissue of the three grasses.
Next steps: Knowledge of the cold-responsive regulatory mechanism will facilitate the engineering of strong cold tolerance in cereal crops via genetic modification of the corresponding regulatory elements. The DHS maps generated in this study provide a resource for identifying cold-responsive regulatory elements, including the most challenging to identify tissue-specific regulatory elements, which may be located far from the target genes. The putative regulatory network generated in this study could facilitate the discovery of cold-responsive regulatory mechanisms in plants.
Han et al. (2020). Genome-wide Characterization of DNase I-hypersensitive Sites and Cold Response Regulatory Landscapes in Grasses. Plant Cell; DOI: https://doi.org/10.1105/tpc.19.00716