Camelina: A History of Polyploidy, Chromosome Shattering, and Recovery
 Following formation of a polyploid plant, cells need to manage issues such as gene dosage and chromosome pairing/segregation to help the formerly separate genomes get along in the same nucleus. As the genomes adjust, chromosomes may undergo recombination, rearrangements, gene loss, and other perturbations, some severe. In some cases, these perturbations mean extinction, but in other (very rare) cases, the cell’s machinery can rebuild the chromosomes. Over time, the polyploid may revert back to a diploid-like genome, as duplicated genes are lost (termed fractionation) or diverge in function (reviewed in Schranz et al., 2006).
Following formation of a polyploid plant, cells need to manage issues such as gene dosage and chromosome pairing/segregation to help the formerly separate genomes get along in the same nucleus. As the genomes adjust, chromosomes may undergo recombination, rearrangements, gene loss, and other perturbations, some severe. In some cases, these perturbations mean extinction, but in other (very rare) cases, the cell’s machinery can rebuild the chromosomes. Over time, the polyploid may revert back to a diploid-like genome, as duplicated genes are lost (termed fractionation) or diverge in function (reviewed in Schranz et al., 2006).
The set of species with different ploidies that evolved from the hybrids and the parental diploids, constitute a polyploid complex. For example, the genus Camelina, a relative of Arabidopsis and Brassica species, has diploid, tetraploid, and hexaploid species. The hexaploid species Camelina sativa (also known as false flax and gold-of-pleasure) may be the next up-and-coming oilseed crop (Vollmann and Eynck, 2015)—in fact, up-and-coming-back might be a better term, as C. sativa has been cultivated for thousands of years but lost favor to other oilseed crops such as canola (Brassica sp.). Now the low-input cultivation and oil properties of C. sativa have attracted renewed interest. For example, high long-chain hydrocarbon contents make camelina oil potentially suitable for use in aviation, and high omega-3-fatty acid contents make it an attractive dietary oil. However, Camelina has been improved only a little by breeding and its origins remain a mystery.
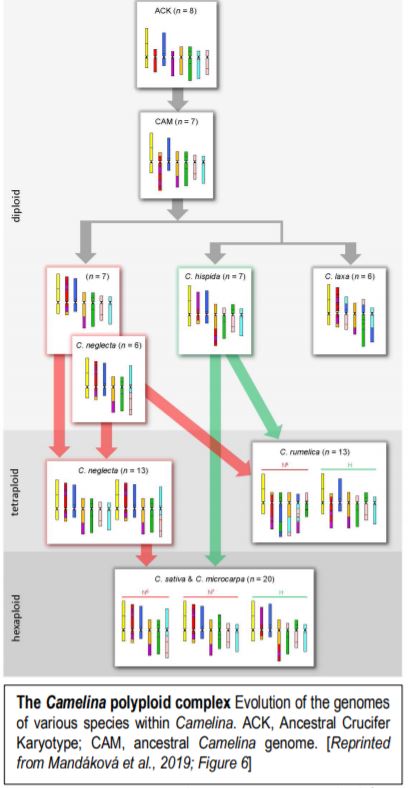
Now, Mandáková et al., (2019) have examined genome evolution in the Camelina polyploid complex. To this end, they first used chromosome-specific contigs of Arabidopsis thaliana bacterial artificial chromosomes to examine how the chromosomes in Camelina diploids relate to the previously inferred Ancestral Crucifer Karyotype (ACK, n=8) (Schranz et al., 2006). Examination of three diploid Camelina species allowed the authors to define the ancestral Camelina genome (CAM, n=7). The CAM evolved from the ACK through recombination between three chromosomes, reducing the chromosome number to seven (figure). The authors then used the CAM to infer how the genomes of the diploids formed, via translocations and other rearrangements that, in some cases, further reduced the chromosome number from 7 to 6, a changed termed descending dysploidy.
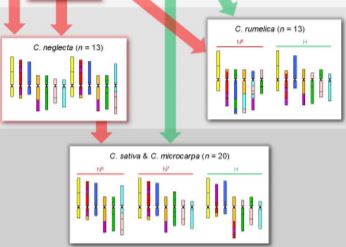
Having established the CAM and diploid chromosome structures, the authors next move up in complexity, examining the allotetraploid (n=13) Camelina rumelica. They use genomic in situ hybridization (GISH) with genomic DNA from the diploids to identify the most probable parental species (with the usual caveat that the existing diploid species are actually the descendants of the parents) and assign the C. rumelica chromosomes to the appropriate parental diploid genomes. Intriguingly, one C. rumelica subgenome has not remained stable following polyploid formation, but has undergone major rearrangements (fractionation).
Moving upward in ploidy, the authors next examine the hexaploids C. sativa and its wild relative, C. microcarpa. GISH results suggested that these hexaploids originated from a tetraploid (extinct or yet to be identified) with a genome similar to that of the diploid C. neglecta, and a diploid similar to C. hispida. Notably, the subgenomes within these hexaploids have stayed stable and not undergone large translocations between homoeologs. The authors further conduct a phylogenetic analysis of a set of 48 single-copy, nuclear genes to support the inferred origins of these species.
This study explores the repeated hybridization and subgenome stasis vs. rearrangements that occurred over the history of this polyploid complex. Moreover, it shows that the descending dysploidies in the diploids involved complex chromosomal rearrangements with many breakpoints, termed chromosome shattering. Understanding the origin of the C. sativa genome sets the stage for improvements of this potentially important oilseed crop, possibly through metabolic engineering in the diploids (which should be more amenable to various manipulations) followed by production of a new hexaploid.
Science Editor
Jennifer Mach, [email protected]
http://orcid.org/0000-0002-1141-6306
REFERENCES
Mandáková, T., Pouch, M., Brock, J.R., Al-Shehbaz, I.A., and Lysak, M.A. (2019). Origin and Evolution of Diploid and Allopolyploid Camelina Genomes was Accompanied by Chromosome Shattering Plant Cell DOI: https://doi.org/10.1105/tpc.19.00366.
Schranz ME, Lysak MA, Mitchell-Olds T. (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006 Nov;11(11):535-42.
Vollmann, J., and Eynck, C. (2015) Camelina as a sustainable oilseed crop: Contributions of plant breeding and genetic engineering. Plant Biotechnolgy Journal https://doi.org/10.1002/biot.201400200