An Emerging Model Diatom to Study Nitrogen Metabolism
Carbon and nitrogen metabolism are intricately linked in all organisms and are tightly regulated to maintain growth, homeostasis, and other cellular activities. In plants and algae, photosynthesis provides both carbon skeletons and the reductant needed for assimilation of inorganic NO3¯ by nitrate reductase (NR). NR catalyzes the conversion of nitrate (NO3¯) to nitrite (NO2¯), which can be further reduced to NH4+ for incorporation into amino acids and other biological molecules. In addition to being potential sources of biofuels and other high value products, marine algae are responsible for as much as 20% of global primary production.
The marine diatom Phaeodactylum tricornutum is an emerging model system for metabolic studies. Cells can be grown asexually in continuous culture with either glycerol or CO2 as a carbon source and can use either NH4+ or NO3¯ as a nitrogen (N) source. It has a relatively small sequenced genome (Bowler et al., 2008) and there are tools for transformation, overexpression, reverse genetics, and RNAi gene silencing (Siaut, et al., 2007; De Riso et al., 2009). Also available are RNAseq libraries, small RNA data and a collection of 130,000 ESTs (Maheswari, et al., 2009). In addition, P. tricornutum produces up to 30% of its dry cell weight in lipids, making it a likely prospect for biofuel production.
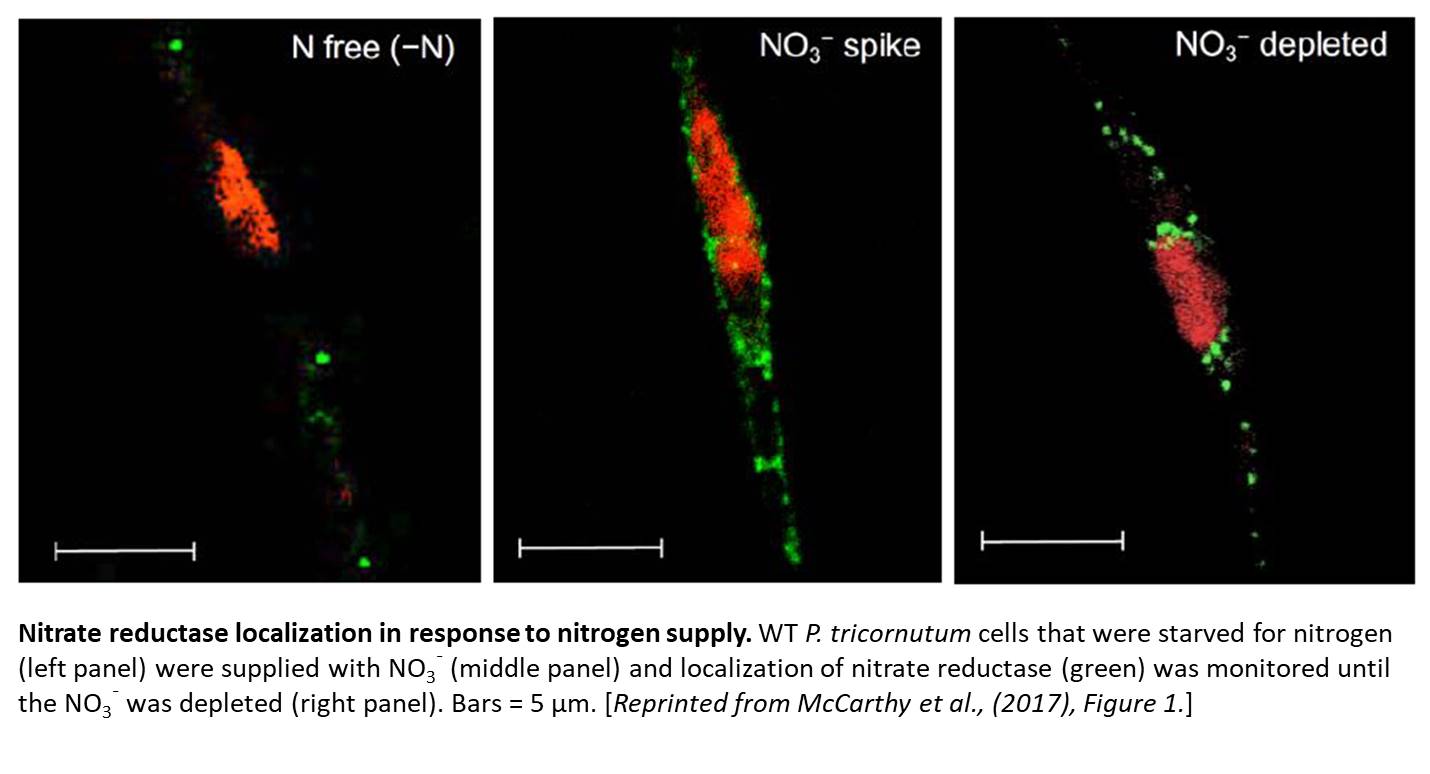
McCarthy et al. () overexpressed a YFP-NR fusion in wild-type (WT) cells and showed that NR was localization in cells was dependent on N availability. In NO3¯-replete conditions, YFP-NR was found throughout the cytoplasm, but during stationary phase, as N becomes limiting, the YFP-NR aggregated into punctate dots. Similarly, when grown on NH4+, which represses NO3¯ uptake and NR activity, native NR appeared as punctate dots. But when NO3¯ was added to N-limited cells, immunolabeled NR was found throughout the cytoplasm until N was depleted (see figure). By coexpressing a fusion of CPF with a known peroxisomal marker, the authors show that the protein was cytosolic until N became limiting and then moved to the peroxisome as extracellular NO3¯ was depleted.
A P. tricornutum NR knockout (NR-KO) had no detectable NR and could not grow on NO3¯ as a sole carbon source. Both defects were complemented using the native NR promoter driving a YFP-NR fusion. Following growth on NH4+ and transfer to NO3¯, both WT and NR-KO rapidly imported NO3¯. Whereas WT cells assimilated all of the internalized NO3¯, NR-KO cells continued to import NO3¯ long after growth had ceased, presumably due to lack of NR-dependent NO3¯ assimilation. After several days, the NR-KO cells became swollen with enlarged vacuoles that presumably contained the unassimilated NO3¯.
A cohort of 35 genes coexpressed with NR, including genes encoding NO3¯ and NO2¯ transporters, chloroplast NR, and ornithine/urea cycle enzymes. Many of these cohort genes were induced to 2 to 4 times WT levels. In addition, the loss of NR upregulated of genes involved in fatty acid, sphingolipid, and triacylglycerol synthesis and caused accumulation of lipid bodies. By contrast, genes involved in light harvesting, photosynthetic electron transport, chlorophyll biosynthesis, and the Calvin cycle were down regulated in NR-KO.
The authors propose a compelling physiological and transcriptomic model in which NR-KO cells exposed to NO3¯ respond simultaneously to signals for both NO3¯-replete and NO3¯ deficient conditions. They respond to the availability of NO3¯ as do WT cells but, because they cannot assimilate this form of nitrogen, they simultaneously experience N-deficient conditions. The NR-KO mutant should benefit not only basic research to understand NO3¯ assimilation and its link to lipid synthesis but also applied research to maximize algal biofuel production.
REFERENCES
Maheswari, U., Mock, T., Armbrust, E.V., Bowler, C. 2009. Update of the Diatom EST Database: a new tool for digital transcriptomics. Nucleic Acids Research 37: D1001-1005.
Bowler, C., et al. (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239-244.
McCarthy, J., et al. (2017). Nitrate Reductase Knockout Uncouples Nitrate Transport from Nitrate Assimilation and Drives Repartitioning of Carbon Flux in a Model Pennate Diatom. Plant Cell 29: doi: 10.1105/tpc.15.00400.
Siaut, M., Heijde, M., Mangogna, M., Montsant, A., Coesel, S., Allen, A., Manfredonia, A., Falciatore, A., and Bowler, C. (2007). Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406: 23-35.
De Riso, V., Raniello, R., Maumus, F., Rogato, A., Bowler, C., Falciatore, A. (2009). Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Research, 37: 4.



Leave a Reply
Want to join the discussion?Feel free to contribute!