Bacterial pathogens deliver water- and solute-permeable channels to plant cells
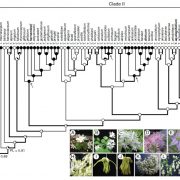
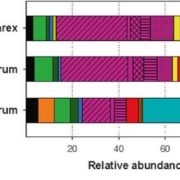
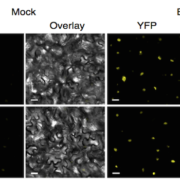
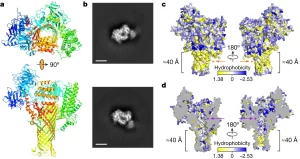 What do you do when you’ve identified a gene that you know is important, but you don’t know how it functions? Usually, you can get hints from homology searches, overexpression studies, or the identification of protein domains, but sometimes those approaches don’t work. That’s where the story of the pathogen virulence protein AvrE from Psueodomonas syringae (and related proteins) sat for a long time. Now, Nomura et al. have cracked the case. They started with AlphaFold2 analysis, the AI system that predicts protein structures. Interestingly, the structural model predicted a “mushroom shaped” protein with a clear central channel. The authors then used several methods to test this model, including cryoEM imaging. Expression in oocytes revealed that the protein creates a functional channel for water and small molecules. Further studies suggested that the channels could be blocked by polyamidoamine dendrimers, and, interestingly, they authors found that these molecules also inhibit the virulence of the pathogens. This exciting paper not only solves a decades-old riddle, but also beautifully illustrates how the combination of AlphaFold2 and cryoEM (both recent advancements) can provide clues to protein function. (Summary by Mary Williams @PlantTeaching) Nature 10.1038/s41586-023-06531-5
What do you do when you’ve identified a gene that you know is important, but you don’t know how it functions? Usually, you can get hints from homology searches, overexpression studies, or the identification of protein domains, but sometimes those approaches don’t work. That’s where the story of the pathogen virulence protein AvrE from Psueodomonas syringae (and related proteins) sat for a long time. Now, Nomura et al. have cracked the case. They started with AlphaFold2 analysis, the AI system that predicts protein structures. Interestingly, the structural model predicted a “mushroom shaped” protein with a clear central channel. The authors then used several methods to test this model, including cryoEM imaging. Expression in oocytes revealed that the protein creates a functional channel for water and small molecules. Further studies suggested that the channels could be blocked by polyamidoamine dendrimers, and, interestingly, they authors found that these molecules also inhibit the virulence of the pathogens. This exciting paper not only solves a decades-old riddle, but also beautifully illustrates how the combination of AlphaFold2 and cryoEM (both recent advancements) can provide clues to protein function. (Summary by Mary Williams @PlantTeaching) Nature 10.1038/s41586-023-06531-5