Autoregulation of RCO by low-affinity binding modulates cytokinin action and shapes leaf diversity (Curr. Biol)
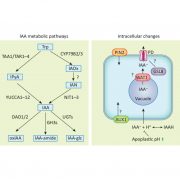
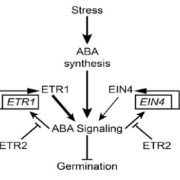
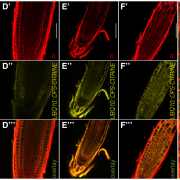
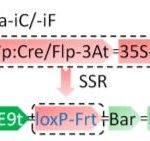
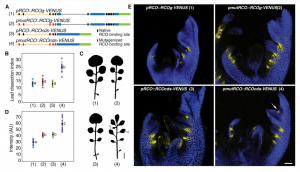 REDUCED COMPLEXITY (RCO) is the main homeodomain transcription factor (TF) responsible for leaf complexity in cruciferous plants. RCO is lost in the model Arabidopsis with simple leaves but present in the genome of Cardamine hirsuta, a good model for leaf morphology traits. However, little is known about RCO’s genome-wide targets. In this paper, Hajheidari et al. address this question using ChiP-seq and RNA-seq in C. hirsuta. Among the identified targets, the authors found that RCO regulates cytokinin signaling, activating the expression of genes involved in CK homeostasis and signaling and locally increasing CK activity in the leaflets. Additionally, RCO expression is autoregulated by repressing itself. This observation is consistent with gene expression analysis and it is crucial to the precise expression of the gene in the leaflets. Instead of a single DNA binding site in the promoter region, the authors found 11 non-canonical low-affinity binding motifs along a distal enhancer and intronic region that mediating the negative feedback-loop and contribute to the fine-tuning of the gene expression. Low-affinity binding sites put in question the simplified logic of transcriptional regulation and can coexist with other regulatory modes in the context of different loci. This kind of interaction was also described for other homeodomain TFs in animals. (Summary by Facundo Romani) Curr. Biol. 10.1016/j.cub.2019.10.040
REDUCED COMPLEXITY (RCO) is the main homeodomain transcription factor (TF) responsible for leaf complexity in cruciferous plants. RCO is lost in the model Arabidopsis with simple leaves but present in the genome of Cardamine hirsuta, a good model for leaf morphology traits. However, little is known about RCO’s genome-wide targets. In this paper, Hajheidari et al. address this question using ChiP-seq and RNA-seq in C. hirsuta. Among the identified targets, the authors found that RCO regulates cytokinin signaling, activating the expression of genes involved in CK homeostasis and signaling and locally increasing CK activity in the leaflets. Additionally, RCO expression is autoregulated by repressing itself. This observation is consistent with gene expression analysis and it is crucial to the precise expression of the gene in the leaflets. Instead of a single DNA binding site in the promoter region, the authors found 11 non-canonical low-affinity binding motifs along a distal enhancer and intronic region that mediating the negative feedback-loop and contribute to the fine-tuning of the gene expression. Low-affinity binding sites put in question the simplified logic of transcriptional regulation and can coexist with other regulatory modes in the context of different loci. This kind of interaction was also described for other homeodomain TFs in animals. (Summary by Facundo Romani) Curr. Biol. 10.1016/j.cub.2019.10.040