AT the Onset of DNA Replication in Arabidopsis
Author
Anne-Sophie Fiorucci
Affiliation
Centre for Integrative Genomics, Faculty of Biology and Medicine, Génopode Building, University of Lausanne, CH-1015 Lausanne, Switzerland.
ORCID number
0000-0002-3254-5967
DNA replication allows doubling of the genomic content of a cell before division. In eukaryotes, it starts at multiple places called origins of replication (ORI) where the replication machinery is loaded (Leonard and Mechali, 2013). The activation timing follows a highly regulated program, with some ORIs activated early and others late during S phase (Concia et al., 2018). However, only 20 to 30% of ORIs are initiated during a given cell cycle and it is still not clear how they are chosen (Ganier et al., 2019).
Most of what we know about DNA replication initiation in eukaryotes comes from studies in yeast and metazoans (Ganier et al., 2019). The main actors of the replication machinery are conserved in plants (Pedroza-Garcia et al., 2019) but plants lack several factors that are required for DNA replication in metazoans. Features of origins of replication also tend to vary a lot between organisms, which means that genome-wide studies are required to better understand the regulation of the replication process in plants. A common protocol to identify ORIs is the analysis of short nascent strands (SNS), a technique that selects and characterizes newly synthesized DNA strands where replication has begun. This method has been widely used in animal models and in plants, using synchronized cells in culture (Costas et al., 2011; Vergara et al., 2017) or more recently entire seedlings at two stages of development (Sequeira-Mendes et al., 2019). However, in all tested systems, the SNS method has shown some limitations, like the potential contamination by non-nascent DNA and difficulties obtaining reproducible results (Hyrien, 2015). Furthermore, techniques to synchronize cells in culture like sucrose starvation are quite stressful for cells and can potentially impact the choice of ORIs.
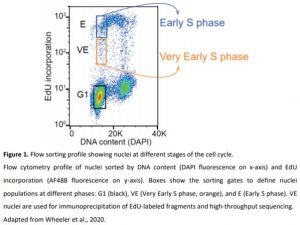 In this issue of Plant Physiology, Wheeler et al. (2020) describe a new method to identify the genomic regions where DNA replication starts in Arabidopsis thaliana (Wheeler et al., 2020). The authors isolated nuclei from wild-type cultured cells and sorted them according to their DNA content and the incorporation of a thymidine analog (EdU) that can be detected after adding a fluorescent dye. To identify the loci where replication is initiated, the authors focused on nuclei that have incorporated a small amount of EdU without much increase in DNA content. These nuclei are considered to be in very early S phase (Figure 1). Indeed, one can assume that replication has just started in these nuclei and that EdU-labeled DNA fragments will be representative of potential replication initiation loci. After immunoprecipitation, high-throughput sequencing of these fragments further allowed the authors to identify the genomic regions located close to the ORIs. The authors called these regions initiation regions (IR), and not ORIs, because the approach does not have enough resolution to locate events at a precise nucleotide. The hypothesis is that IRs are not ORIs per se but must contain or be located near ORIs.
In this issue of Plant Physiology, Wheeler et al. (2020) describe a new method to identify the genomic regions where DNA replication starts in Arabidopsis thaliana (Wheeler et al., 2020). The authors isolated nuclei from wild-type cultured cells and sorted them according to their DNA content and the incorporation of a thymidine analog (EdU) that can be detected after adding a fluorescent dye. To identify the loci where replication is initiated, the authors focused on nuclei that have incorporated a small amount of EdU without much increase in DNA content. These nuclei are considered to be in very early S phase (Figure 1). Indeed, one can assume that replication has just started in these nuclei and that EdU-labeled DNA fragments will be representative of potential replication initiation loci. After immunoprecipitation, high-throughput sequencing of these fragments further allowed the authors to identify the genomic regions located close to the ORIs. The authors called these regions initiation regions (IR), and not ORIs, because the approach does not have enough resolution to locate events at a precise nucleotide. The hypothesis is that IRs are not ORIs per se but must contain or be located near ORIs.
By comparing several genomic features (GC content, chromatin context, gene/transposable element presence), the authors characterized IRs as AT-rich sequences mostly associated with intergenic regions and flanked by relatively GC-rich sequences. They further classified IRs into two categories based on the strength of the EdU signal after immunoprecipitation. Strong IRs are located regularly along the chromosome arms whereas weak IRs are preferentially found in pericentromeric regions. To examine a potential link with chromatin accessibility, the authors also treated the samples with micrococcal nuclease and observed that IRs are generally associated with open chromatin regions.
The observation by Wheeler et al. (2020) that IRs are associated with AT-rich regions of the genome contrasts with previous reports on Arabidopsis cells or seedlings. Other studies indeed located ORIs in the vicinity of GC-rich sequences mostly associated either with the 5’ end of genes in the chromosome arms (Costas et al., 2011; Sequeira-Mendes et al., 2019) or with transposable elements in pericentromeric regions (Vergara et al., 2017). These results are consistent with what is described for ORIs in metazoans (Ganier et al., 2019). However, similar to the results presented by Wheeler et al., AT-rich regions are used as ORIs in other organisms like yeast (Schizosaccharomyces pombe for example). In human cells, some ORIs that are kept dormant (not activated for replication) are also associated with GC-poor regions (Ganier et al., 2019). Results of the different studies in plants might therefore not be mutually exclusive but they might actually describe different types of ORIs with specific functions, in different plant tissues or cells under different conditions. The observed inconsistencies might illustrate the differences between the Landsberg erecta (Costas et al., 2011; Vergara et al., 2017) and Col-0 ecotypes (Wheeler et al., 2020), or between cells in culture and entire seedlings (Sequeira-Mendes et al., 2019). The use of different techniques to identify ORIs (SNS versus the new method described in Wheeler et al.), each having their own bias, might also explain part of the discrepancies.
Even if DNA replication looks like a well-known textbook process, the paper by Wheeler et al. (2020) provides new insights on replication origins in plants and surely opens the way for future exciting understandings of the regulation of a major cellular process.
References
Concia, L., Brooks, A.M., Wheeler, E., Zynda, G.J., Wear, E.E., LeBlanc, C., Song, J., Lee, T.J., Pascuzzi, P.E., Martienssen, R.A., Vaughn, M.W., Thompson, W.F., and Hanley-Bowdoin, L. (2018). Genome-Wide Analysis of the Arabidopsis Replication Timing Program. Plant Physiol 176, 2166-2185.
Costas, C., de la Paz Sanchez, M., Stroud, H., Yu, Y., Oliveros, J.C., Feng, S., Benguria, A., Lopez-Vidriero, I., Zhang, X., Solano, R., Jacobsen, S.E., and Gutierrez, C. (2011). Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks. Nat Struct Mol Biol 18, 395-400.
Ganier, O., Prorok, P., Akerman, I., and Mechali, M. (2019). Metazoan DNA replication origins. Curr Opin Cell Biol 58, 134-141.
Hyrien, O. (2015). Peaks cloaked in the mist: the landscape of mammalian replication origins. J Cell Biol 208, 147-160.
Leonard, A.C., and Mechali, M. (2013). DNA replication origins. Cold Spring Harb Perspect Biol 5, a010116.
Pedroza-Garcia, J.A., De Veylder, L., and Raynaud, C. (2019). Plant DNA Polymerases. Int J Mol Sci 20.
Sequeira-Mendes, J., Vergara, Z., Peiro, R., Morata, J., Araguez, I., Costas, C., Mendez-Giraldez, R., Casacuberta, J.M., Bastolla, U., and Gutierrez, C. (2019). Differences in firing efficiency, chromatin, and transcription underlie the developmental plasticity of the Arabidopsis DNA replication origins. Genome Res 29, 784-797.
Vergara, Z., Sequeira-Mendes, J., Morata, J., Peiro, R., Henaff, E., Costas, C., Casacuberta, J.M., and Gutierrez, C. (2017). Retrotransposons are specified as DNA replication origins in the gene-poor regions of Arabidopsis heterochromatin. Nucleic Acids Res 45, 8358-8368.
Wheeler, E., Brooks, A.M., Concia, L., Vera, D., Wear, E.E., LeBlanc, C., Ramu, U., Vaughn, M.W., Bass, H., Martienssen, R., Thompson, W.F., and Hanley-Bowdoin, L. (2020). Arabidopsis DNA replication initiates in intergenic, AT-rich open chromatin. Plant Physiol. https://doi.org/10.1104/pp.19.01520