ASTREL Projection: Comparative Phylogenomics Uncovers Novel Genes Co-eliminated with the EDS1 Immune Pathway
The plant immune system is largely centered around immune receptors that monitor for pathogen-derived signatures. Extracellular immunity is afforded through cell-surface receptors recognizing common microbial motifs like bacterial flagellin or fungal chitin (Boutrot and Zipfel, 2017). By contrast, intracellular immunity relies on disease resistance (R) receptors that directly or indirectly perceive pathogen virulence effectors attempting to subvert host defenses. Importantly, these receptor proteins are present across the green plant lineage and typically display a conserved/canonical “NLR” structure that includes a variable N-terminus followed by the NB-ARC domain and leucine-rich repeats (van Wersch et al., 2020).
Decades of research has solidified the role of NLRs as key mediators of effector-triggered immunity (ETI) in a limited number of flowering plants (angiosperms). Despite overwhelming functional evidence in crops and genetic model systems, we currently lack a greater understanding of the evolutionary dynamics driving NLR-mediated immunity across distantly-related plants. To gain new evolutionary insights into NLR-mediated immune mechanisms, Baggs et al. (2020) took a comparative phylogenomic approach to characterize NLRs and NLR-associated immune machinery present across diverse plant species.
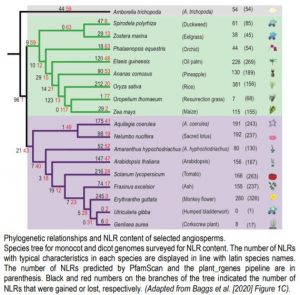 Using publicly available genome data, the authors surveyed the NLR repertoires of 95 angiosperms spanning 24 distinct orders. While the majority of analyzed species encode between 200 and 500 NLRs, extensive gains and losses of NLR content were occasionally observed. In particular, gene loss events occurring in eel grass (Zostera marina), resurrection grass (Oropetium thomaeum), greater duckweed (Spirodela polyrhiza), orchid (Phalaenopsis equestris), maize (Zea mays), pineapple (Ananas comosus), humped bladderwort (Utricularia gibba), and corkscrew plant (Genlisea aurea) resulted in smaller than normal NLR sets compared to other angiosperms (Figure). Phylogenetic analysis based on the conserved NB-ARC domain further illustrated extensive losses and gains of NLRs in certain lineages, with the loss of NLRs independently occurring in the ancestor of Z. mays and O. thomaeum, and in the ancestor of U. gibba and G. aeurea.
Using publicly available genome data, the authors surveyed the NLR repertoires of 95 angiosperms spanning 24 distinct orders. While the majority of analyzed species encode between 200 and 500 NLRs, extensive gains and losses of NLR content were occasionally observed. In particular, gene loss events occurring in eel grass (Zostera marina), resurrection grass (Oropetium thomaeum), greater duckweed (Spirodela polyrhiza), orchid (Phalaenopsis equestris), maize (Zea mays), pineapple (Ananas comosus), humped bladderwort (Utricularia gibba), and corkscrew plant (Genlisea aurea) resulted in smaller than normal NLR sets compared to other angiosperms (Figure). Phylogenetic analysis based on the conserved NB-ARC domain further illustrated extensive losses and gains of NLRs in certain lineages, with the loss of NLRs independently occurring in the ancestor of Z. mays and O. thomaeum, and in the ancestor of U. gibba and G. aeurea.
The convergent loss of NLRs led the authors to speculate that other immune components are likely to be lost alongside NLRs. Genome-wide surveys revealed a clear loss of the EDS1 (ENHANCED DISEASE SUSCEPTIBILITY1) immune branch and NDR1 (NON-HOST DISEASE RESISTANCE1) in angiosperms with extensively reduced NLR content. Taking this concept further, the authors predicted that otherwise unknown components of the EDS1-dependent NLR immune pathway are similarly co-eliminated alongside EDS1/NLRs in these plants (S. polyrhiza, Z. marina, G. aurea, U. gibba, and Asparagus officinalis). Using orthology-based analyses, Baggs and colleagues identified a set of ASTREL (AngioSperm Typically Retained, EDS1-Lost) genes, representing new candidates with potential roles in EDS1-dependent immunity.
Lastly, co-expression analysis of public gene expression data was performed to gain initial insights into ASTREL gene regulation in NLR-typical model plants. Intriguingly, this analysis linked differential expression during pathogen infection with drought stress or ABA accumulation, which is particularly exciting given that many of the angiosperms lacking EDS1/ASTREL genes are either fully or partially aquatic with constant exposure to water. Future analyses may yet reveal key links between water status and EDS1-dependent NLR immunity.
Philip Carella
Sainsbury Laboratory
University of Cambridge, Cambridge
ORCID: 0000-0002-5467-7290
REFERENCES
Baggs EL, Monroe JG, Thanki AS, O’Grady R, Schudoma C, Haerty W and Krasileva KV (2020). Convergent loss of an EDS1/PAD4 signaling pathway in several plant lineages reveals co-evolved components of plant immunity and drought response. Plant Cell Published May 2020. DOI: https://doi.org/10.1105/tpc.19.00903.
Boutrot F and Zipfel C (2017). Function, Discovery, and Exploitation of Plant Pattern Recognition Receptors for Broad-Spectrum Disease Resistance. Annu Rev Phytopathol 55: 257–286.
van Wersch S, Tian L, Hoy R and Li X (2020). Plant NLRs: The Whistleblowers of Plant Immunity. Plant Communications 1: 100016.



