Assembling a Nanomolecular Power Station
The ATP synthase complex of chloroplasts is an elegant example of the union of structure and function at the molecular level (Junge and Nelson, 2015). This enzyme complex consists of an integral membrane CFo component that transports protons and an extrinsic CF1 component that synthesizes ATP (Hahn et al., 2018). CFo uses the potential energy of a light-induced proton gradient to spin a ring of c subunits in the membrane. Via a connecting γ subunit, the c ring acts as a nanomotor and transfers the energy to the extrinsic catalytic α3β3 subunits of CF1, resulting in ATP production.
Five different types of subunits (α, β, γ, δ, and ε) assemble precisely to form the CF1 component of the mature CF1CFo ATP synthase, but the assembly mechanism is poorly understood. An extensive screen had identified Arabidopsis mutants showing the high level of nonphotochemical quenching that is typical of ATP synthase mutants (Zhang, et al. 2016). These bfa mutants (BIOGENESIS FACTOR REQUIRED FOR ATP SYNTHASE) are deficient in the assembly of the chloroplast ATP synthase. Here, Zhang, et al. (2018) characterize the bfa1 mutant in detail, report the crystal structure of BFA1, and analyze BFA1 interactions with each of the five CF1 subunits. Based on molecular docking, yeast two-hybrid, pull-down, and coimmunoprecipitation analyses, they propose a very nice model for how BFA1 interacts with CF1 subunits β and γ to promote the assembly of functional CF1 in Arabidopsis.
Levels of all five CF1 subunits were specifically reduced in bfa1, whereas subunit levels of several other thylakoid protein complexes were unaffected. In addition, bfa1 had a lower electron transport rate through photosystem II and a higher reduction state of the plastoquinone pool, consistent with increased accumulation of protons in the thylakoid lumen due to ATP synthase deficiency.
 Pulse-chase experiments showed that newly synthesized CF1α and CF1β subunits were barely detectable in the mature bfa1 ATP synthase in contrast to the wild type, confirming the role of BFA1 in CF1 assembly. Using map-based cloning, the authors localized BFA1 to chromosome 3, and identified it as At3g29185, which encodes a protein of previously unknown function. The bfa1 mutant did not produce the At3g29185 transcript, and the bfa1 mutation was fully complemented by the wild-type At3g29185 sequence. BFA1 was localized in the chloroplast stroma of wild-type plants, but was absent from bfa1 mutant plants.
Pulse-chase experiments showed that newly synthesized CF1α and CF1β subunits were barely detectable in the mature bfa1 ATP synthase in contrast to the wild type, confirming the role of BFA1 in CF1 assembly. Using map-based cloning, the authors localized BFA1 to chromosome 3, and identified it as At3g29185, which encodes a protein of previously unknown function. The bfa1 mutant did not produce the At3g29185 transcript, and the bfa1 mutation was fully complemented by the wild-type At3g29185 sequence. BFA1 was localized in the chloroplast stroma of wild-type plants, but was absent from bfa1 mutant plants.
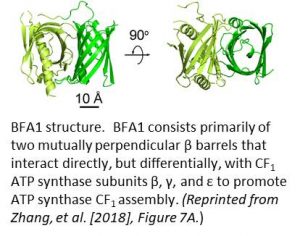
The authors solved the crystal structure of BFA1 at 2.8 Å resolution and found that it is composed primarily of two β barrels oriented at right angles to each other (see figure). Yeast two-hybrid studies showed that BFA1 interacted with CF1β, CF1γ, and CF1ε, but not with CF1α and CF1δ. The authors performed a more detailed binding analysis and determined which domains of CF1β and CF1γ bind to which specific β barrels of BFA1. As further confirmation of its role in ATP synthase assembly, an in silico molecular docking analysis showed that both the CF1β and the CF1γ subunits would fit closely on the surface of BFA1 without steric conflicts. Continued studies with bfa mutants will certainly improve our understanding of ATP synthase assembly, and they are likely to provide inspiration in our attempts to develop synthetic molecular motors for nanomedicine.
REFERENCES
Hahn, A., Vonck, J., Mills, D.J., Meier, T., and Kühlbrandt, W. (2018). Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 360: eaat4318. DOI: 10.1126/science.aat4318.
Junge, W. and Nelson, N. (2015). ATP Synthase. Annu. Rev. Biochem. 84: 631–657.
Zhang, L., Duan, Z., Zhang, J., and Peng, L. (2016). BIOGENESIS FACTOR REQUIRED FOR ATP SYNTHASE 3 facilitates assembly of the chloroplast ATP synthase complex. Plant Physiol. 171: 1291–1306.
Zhang, L., Pu, H., Duan, Z., Li, Y., Liu, B., Zhang, Q., Li, W., Rochaix, J-D., Liu, L., and Peng, L. (2018). Nucleus-encoded Protein BFA1 Promotes Efficient Assembly of the Chloroplast ATP Synthase Coupling Factor 1. Plant Cell. DOI: 10.1105/tpc.18.00075.