Arabidopsis and crop plants differ in their 3D genome architecture ($)
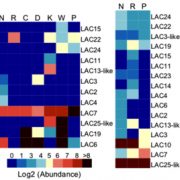
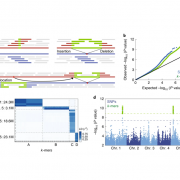
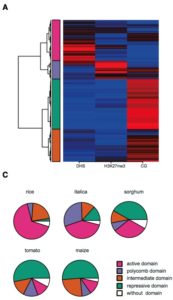 Three-dimensional organization of the genome is critical for proper gene expression. Comparison of the extremely compact genome of Arabidopsis with mammalian genomes revealed reduced local intra-chromosomal contacts (also know as Topologically Associated Domains, TADs) and fewer chromatin loops in the plant model. However, very little is known about spatial chromatin organization in plants with bigger and more complex genomes. In this study, Dong et al. provide the genome 3D architecture of five crop species (rice, sorghum, foxtail millet, tomato and maize). They identified in all five species large-scale euchromatic and heterochromatic compartments that can further be divided into local compartments based on their chromatin marks signature. Contrary to Arabidopsis and similar to mammals, crops exhibit obvious TAD-like domains, suggesting that local contacts are more frequent in large plant genomes. This is associated with an increase in long-range chromatin interactions through loops in the large tomato and maize genomes. This study highlights the complexity and the variety of 3D chromatin architecture in plants, and provides useful genomic and epigenomic resources for the community. (Summary by Matthias Benoit) Mol. Plant 10.1016/j.molp.2017.11.005
Three-dimensional organization of the genome is critical for proper gene expression. Comparison of the extremely compact genome of Arabidopsis with mammalian genomes revealed reduced local intra-chromosomal contacts (also know as Topologically Associated Domains, TADs) and fewer chromatin loops in the plant model. However, very little is known about spatial chromatin organization in plants with bigger and more complex genomes. In this study, Dong et al. provide the genome 3D architecture of five crop species (rice, sorghum, foxtail millet, tomato and maize). They identified in all five species large-scale euchromatic and heterochromatic compartments that can further be divided into local compartments based on their chromatin marks signature. Contrary to Arabidopsis and similar to mammals, crops exhibit obvious TAD-like domains, suggesting that local contacts are more frequent in large plant genomes. This is associated with an increase in long-range chromatin interactions through loops in the large tomato and maize genomes. This study highlights the complexity and the variety of 3D chromatin architecture in plants, and provides useful genomic and epigenomic resources for the community. (Summary by Matthias Benoit) Mol. Plant 10.1016/j.molp.2017.11.005