An Improved plant toolset for high-throughput recombineering (BioRxiv)
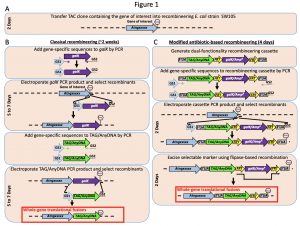
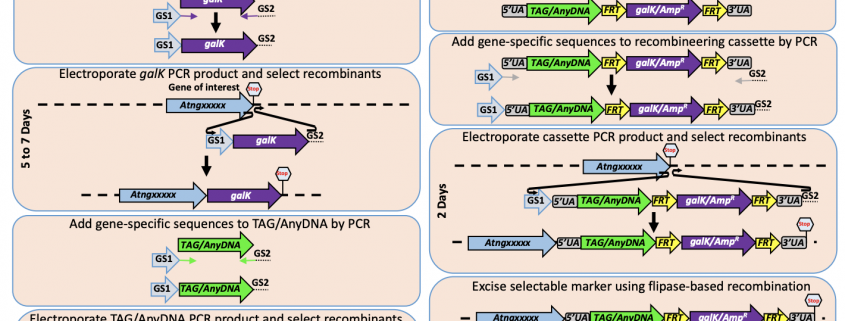
To determine the role of any gene of interest it is necessary to understand the spatiotemporal expression pattern. This is accomplished by tagging the regulatory sequence(s) alone (transcriptional fusion) or regulatory sequence(s) with the coding region (translational fusion) of the gene of interest. In this paper Brumos et al., have developed a recombineering based gene tagging approach that could efficiently tag the gene of interest. In this approach, the authors have created a recombinase-mediated cassette that is available in public stock centers including ABRC, which allows the transfer of sequence from a BAC (Bacterial Artificial Chromosome) clone in to plant transformation compatible binary vector. The efficiency of the system has been evaluated in Arabidopsis by tagging multiple genes involved in auxin-related genes but expandable beyond Arabidopsis. Thus the tool developed here provides more precise gene modifications and reduces the labor/time involved in generating these valuable constructs. (Summarized by Suresh Damodaran) bioRxiv 10.1101/659276