All Together Now: Phylotranscriptomics Reveals Core Responses to Fungal Infection Across the Pentapetalae
Over 450 million years of land plant evolution has led to the acquisition of diverse and often overlapping sets of defense responses that limit pathogen infection. Decades of research has uncovered canonical immune pathways that provide resistance to specialist (hemi)-biotrophic pathogens relying on living plants for nutrition. In comparison, less is known about resistance strategies that protect plants against broad host-range necrotrophic pathogens, which actively destroy host tissues and metabolize the remains (Mbengue et al., 2016).
The necrotrophic fungal pathogen Sclerotinia sclerotiorum causes white mold on a wide range of dicot crops and model systems (Bolton et al., 2006). Unlike qualitative resistance mechanisms typically acting against specialist pathogens, plants infected with Sclerotinia exhibit quantitative disease resistance (QDR), where infection outcomes are continuously distributed and cannot be separated into discrete ‘resistant’ or ‘susceptible’ groups (Corwin and Kliebenstein, 2017). To elucidate evolutionarily-conserved processes contributing to plant QDR, Sucher et al. (2020) surveyed host transcriptional responses to infection by a single S. sclerotiorum isolate in six distantly-related species within the Pentapetalae clade of eudicots. This included individual genotypes of important eudicot crops and/or genetic model systems: Arabidopsis (Arabidopsis thaliana), beet (Beta vulgaris), common sunflower (Helianthus annuus), common bean (Phaseoulus vulgaris), castor bean (Ricinus communis), and tomato (Solanum lycopersicum).
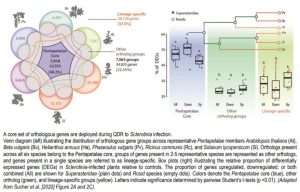 Transcriptome-wide comparisons were made using orthologous gene sets categorized by the presence or absence of genes across each of the six species. The “Pentapetalae core” set included orthologous genes expressed in all six species, “other orthologs” included orthologous genes present in 2-5 species, while “lineage-specific” genes were present in only a single species. Comparative transcriptomics analysis demonstrated that the majority of Sclerotinia infection-responsive genes belonged to the “Pentapetalae core” set of orthologs (Figure). Subsequent evolutionary inferences revealed the progressive acquisition/recruitment of QDR-related genes that are induced during fungal infection in the Pentapetalae.
Transcriptome-wide comparisons were made using orthologous gene sets categorized by the presence or absence of genes across each of the six species. The “Pentapetalae core” set included orthologous genes expressed in all six species, “other orthologs” included orthologous genes present in 2-5 species, while “lineage-specific” genes were present in only a single species. Comparative transcriptomics analysis demonstrated that the majority of Sclerotinia infection-responsive genes belonged to the “Pentapetalae core” set of orthologs (Figure). Subsequent evolutionary inferences revealed the progressive acquisition/recruitment of QDR-related genes that are induced during fungal infection in the Pentapetalae.
To explore the functional relevance of conserved Sclerotinia-responsive genes, the authors performed gene enrichment analyses based on GO (Gene Ontology) and pfam (Protein Family) annotations. Overall, this revealed a limited set of functional annotations enriched with differentially expressed genes in the Pentapetalae core set. Although Sclerotinia-responsive genes tend to be conserved at the interspecific level, their expression upon infection is not. Using simulations for gene expression heritability, the authors suggest that responsiveness to Sclerotinia was acquired vertically only in rare cases. One such exception was the evolutionarily conserved response of a group of ABCG (ATP-BINDING CASETTE G) transporters to Sclerotinia infection across the Pentapetalae. Since Sclerotinia evolved more recently than the radiation of the Pentapetalae, Sucher et al. (2020) reasoned that the Sclerotinia-related expression of ABCG orthologs occurred via exaptation from comparable ancestral stress responses. In support of this, publicly available expression data identified an A. thaliana ABCG40 ortholog consistently induced during bacterial or fungal infection. Subsequent mutant analysis showed a significant role for ABCG40 in QDR to Sclerotinia.
Collectively, the transcriptomic resources and evolutionary insights provided by Sucher et al. (2020) reveal core responses to a necrotrophic fungal infection in the Pentepetalae, which features the upregulation of functionally-relevant ABCG transporters. Future evolutionary and molecular genetic analyses will help to resolve the many genetic mechanisms underpinning QDR in land plants and test whether within and between species variation among hosts and pathogens resamples a core set of mechanisms.
Philip Carella
Sainsbury Laboratory, University of Cambridge, Cambridge
ORCID: 0000-0002-5467-7290
REFERENCES
Bolton MD, Thomma BPHJ, and Nelson BD (2006). Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7: 1–16.
Corwin JA, and Kliebenstein DJ (2017). Quantitative Resistance: More Than Just Perception of a Pathogen. Plant Cell 29: 655–665.
Mbengue M, Navaud O, Peyraud R, Barascud M, Badet T, Vincent R, Barbacci A, and Raffaele S (2016). Emerging Trends in Molecular Interactions between Plants and the Broad Host Range Fungal Pathogens Botrytis cinerea and Sclerotinia sclerotiorum. Front. Plant Sci. 7: 422.