ADP Ribosylation: The Modification Causing a Disease Resistance Sensation
One of the key aspects of pathogenesis is the ability to sabotage host defenses and, to this end, plant pathogens produce a remarkable set of effector proteins that target host defenses at multiple levels. Plants, in turn, have defenses to counteract these effectors; one key aspect of this is the ability to recognize the presence of effectors and initiate defense responses (reviewed in Toruño et al., 2016). This recognition often occurs via perception of a change in the host target of the effector. For example, in Arabidopsis thaliana, the effectors AvrB and AvrRpm1 from the bacterial pathogen Pseudomonas syringae target RPM1-INTERACTING PROTEIN4 (RIN4) for modification. This modification of RIN4 is sensed by the nucleotide binding leucine rich repeat (NLR) protein RESISTANCE TO PSEUDOMONAS MACULICOLA1 (RPM1), which induces defense responses such as programmed cell death, reactive oxygen species production, and callose deposition.
Initial evidence showed that RIN4 underwent phosphorylation in response to AvrRpm1. However, modeling based on protein sequence homology suggested that AvrRpm1 has no similarity to protein kinases—rather, it has similarity to ADP ribosyl transferases (which include, most notably, diphtheria toxin; reviewed in Liu and Wu, 2015). These enzymes transfer an ADP-ribose from nicotinamide adenine dinucleotide (NAD+) to a protein, nucleotide, or other target; they can add a single or multiple ADP-ribose groups, forming a chain. To explore exactly what modifications occurred on RIN4, Redditt et al. (2019) co-expressed soybean (Glycine max) GmRIN4b with AvrRpm1 or AvrB in Nicotiana benthamiana. Indeed, AvrRpm1 (but not AvrB) induced a modification that altered GmRIN4b migration on a gel, but was not sensitive to phosphatase treatment. The modification also occurred in mutant GmRIN4b with substitutions in known phosphorylated amino acids. Immunoprecipitation, followed by mass spectrometry, revealed that GmRIN4b was ADP ribosylated in its N-terminal and C-terminal nitrate-induced (NOI) domains and this was confirmed by immunoblotting with anti-pan-ADP-ribose binding reagent. Further analysis with this reagent showed that AvrRpm1 can ADP ribosylate other RIN4 proteins (including Arabidopsis RIN4) and at least ten Arabidopsis NOI domain-containing proteins. The existence of other targets is consistent with the observation that AvrRpm1 enhances P. syringae virulence, even in rin4 mutants.
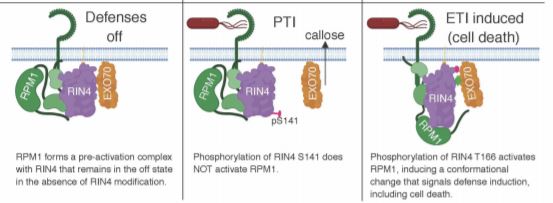
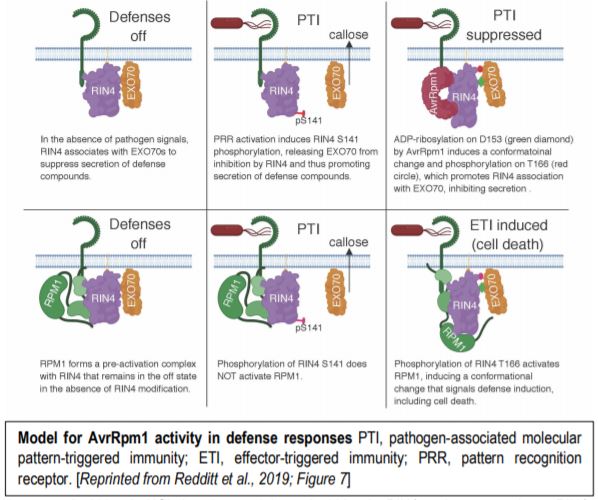 Two things happen in response to RIN4 ADP ribosylation; one is a change in RIN4 activity that benefits the pathogen. AvrRpm1 suppresses callose deposition—callose being a β-1,3-glucan (with a few β-1,6-branches) that plants produce when their cell walls need a little reinforcement—for example, during pollen development, controlling the size exclusion limit in plasmodesmata, as a structural element in xylem sieve tubes, and in cells responding to pathogen attack. Indeed, mutant AvrRpm1 that lacks catalytic activity failed to suppress callose deposition. The exocyst complex functions in secretion of callose and mutants of the EXO70 subunit show reduced callose deposition. Moreover, the RIN4 NOI domain mediates the interaction between Arabidopsis RIN4 and EXO70 exocyst subunits. The RIN4 NOI domain also appears to interact with AvrRpm1, as co-expression of active or catalytically dead AvrRpm1 decreased the co-immunoprecipitation of EXO70 with Arabidopsis RIN4 and some soybean RIN4 proteins. Because overexpression in N. benthamiana appeared to cause competitive binding effects, the authors used yeast two-hybrid to test the effect of modification on the RIN4–EXO70 interaction. Intriguingly, a phosphomimic mutant of RIN4 stabilized the RIN4–EXO70 interaction, consistent with a model in which AvrRpm1-induced modifications increase the ability of RIN4 to suppress callose deposition (figure).
Two things happen in response to RIN4 ADP ribosylation; one is a change in RIN4 activity that benefits the pathogen. AvrRpm1 suppresses callose deposition—callose being a β-1,3-glucan (with a few β-1,6-branches) that plants produce when their cell walls need a little reinforcement—for example, during pollen development, controlling the size exclusion limit in plasmodesmata, as a structural element in xylem sieve tubes, and in cells responding to pathogen attack. Indeed, mutant AvrRpm1 that lacks catalytic activity failed to suppress callose deposition. The exocyst complex functions in secretion of callose and mutants of the EXO70 subunit show reduced callose deposition. Moreover, the RIN4 NOI domain mediates the interaction between Arabidopsis RIN4 and EXO70 exocyst subunits. The RIN4 NOI domain also appears to interact with AvrRpm1, as co-expression of active or catalytically dead AvrRpm1 decreased the co-immunoprecipitation of EXO70 with Arabidopsis RIN4 and some soybean RIN4 proteins. Because overexpression in N. benthamiana appeared to cause competitive binding effects, the authors used yeast two-hybrid to test the effect of modification on the RIN4–EXO70 interaction. Intriguingly, a phosphomimic mutant of RIN4 stabilized the RIN4–EXO70 interaction, consistent with a model in which AvrRpm1-induced modifications increase the ability of RIN4 to suppress callose deposition (figure).
The other response to RIN4 ADP ribosylation, in cells carrying RPM1, is activation of RPM1 and initiation of defense responses. Therefore, the authors explored how ADP ribosylation fits with RIN4 phosphorylation and activation of RPM1. Indeed, analysis in the N. benthamiana system using mutant versions of RIN4 and AvrRpm1 showed that ADP ribosylation promotes RIN4 phosphorylation. Moreover, both modifications are required for RPM1-mediated defense activation. These findings solve the long-standing question of how plant cells perceive AvrRpm1 via effects on RIN4. Exploration of the protein–protein interactions occurring in these systems, including the mechanism by which RIN4 suppresses callose deposition, remains an interesting prospect for future research. Moreover, these findings raise additional intriguing questions—namely, what about those other NOI-domain proteins modified by AvrRpm1? Doubtless, this modification will continue to cause a sensation in research on host–pathogen interactions for some time.
Science Editor
http://orcid.org/0000-0002-1141-6306
REFERENCES
Liu, C., and Yu, X. (2015) ADP-Ribosyltransferases and Poly ADP-Ribosylation. Curr Protein Pept Sci. 2015; 16(6): 491–501.
Reddit, T.J., Chung, E.-H., Karimi, H.Z., Rodibaugh, N., Zhang, Y., Trinidad, J.C., Kim, J.H., Zhou, Q., Shen, M., Dangl, J.L., Mackey, D., and Roger W. Innes, R.W. (2019). AvrRpm1 Functions as an ADP-Ribosyl Transferase to Modify NOI-domain Containing Proteins, Including Arabidopsis and Soybean RPM1-interacting Protein 4. Plant Cell DOI: https://doi.org/10.1105/tpc.19.00020
Toruño TY, Stergiopoulos I, Coaker G. (2016) Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu Rev Phytopathol. 2016 Aug 4;54:419-41. doi: 10.1146/annurev-phyto-080615-100204.