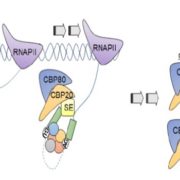
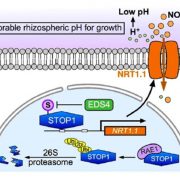
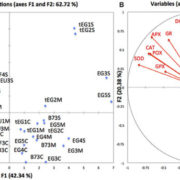
Adaptive diversification of growth allometry in Arabidopsis thaliana (PNAS)
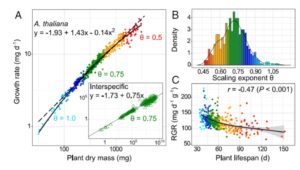 Most students learn that metabolism does not increase at the same rate as an animal’s size – an elephant eats much less per unit of body mouse than a mouse does. This non-linear scaling, described through metabolic scaling theory, also occurs in plants – the growth rate of large plants is relatively slower than that of small plants. The relationship between different facets of growth is known as allometry. To better understand growth allometry in plants, Vasseur et al. examined the relationship between growth rate and final dry mass in 451 Arabidopsis thaliana accessions. They found that although many accessions showed the expected relationship between size and growth rate, some deviated significantly, and that in some cases these deviations (both fast and slow) are correlated with extreme drought tolerance. Analysis of the genetic factors contributing to allometry diversification revealed many genes involved in metabolism and stress responses. They further identifed an important contribution from PUB4, which encodes a U-box protein gene involved in protein turnover, a key regulator of plant responses to abiotic stress. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1709141115
Most students learn that metabolism does not increase at the same rate as an animal’s size – an elephant eats much less per unit of body mouse than a mouse does. This non-linear scaling, described through metabolic scaling theory, also occurs in plants – the growth rate of large plants is relatively slower than that of small plants. The relationship between different facets of growth is known as allometry. To better understand growth allometry in plants, Vasseur et al. examined the relationship between growth rate and final dry mass in 451 Arabidopsis thaliana accessions. They found that although many accessions showed the expected relationship between size and growth rate, some deviated significantly, and that in some cases these deviations (both fast and slow) are correlated with extreme drought tolerance. Analysis of the genetic factors contributing to allometry diversification revealed many genes involved in metabolism and stress responses. They further identifed an important contribution from PUB4, which encodes a U-box protein gene involved in protein turnover, a key regulator of plant responses to abiotic stress. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1709141115