Accuracy matters: What happens when phenotyping errors are non-random?
By Yan Zhou
Iowa State University
Background: The accuracy of trait measurement greatly affects the quality of genetic analyses. During automated phenotyping, trait measurement errors, i.e., differences between automatically extracted trait values and ground truth, are generally treated as random effects that can be controlled by increasing population sizes and/or replication number. There is, however, some evidence that trait measurement errors are partially under genetic control.
Question: Are trait measurement errors at least partially under genetic control? If so, how does this affect genetic analyses?
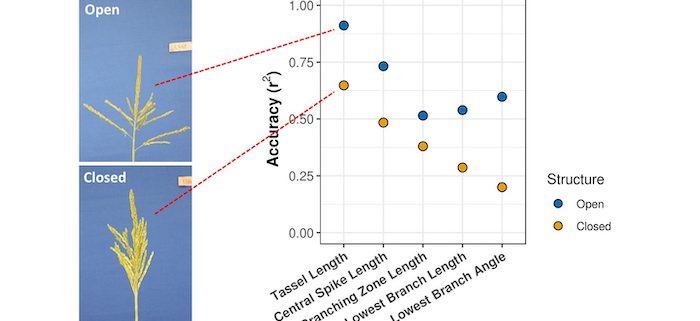
Findings: Consistent with our hypothesis, we observed substantial non-random, genetic contributions to trait measurement errors for five maize tassel traits collected using an image-based phenotyping platform. Phenotyping accuracy varied according to whether a tassel exhibited “open” vs. “closed” branching architecture, which is itself under genetic control. Trait-associated SNPs (TASs) identified via genome-wide association studies (GWASs) conducted on five tassel traits that had been phenotyped both manually (i.e., ground truth) and via feature extraction from images exhibited little overlap. Furthermore, identification of TASs from GWASs conducted on the differences between the two values indicated that a fraction of measurement errors are under genetic control. Such genetically associated measurement errors, which we later defined as genetically determined measurement bias (GDMB), can greatly alter the results of genetic analyses, causing over or under estimation, or even changing the sign, of allelic effects. Consistently, the TASs identified via GWAS on ground truth and automated phenotypic value exhibited little overlap. Similar results were also obtained in a sorghum plant height dataset, demonstrating the existence of GDMB in multiple species and for multiple traits. GDMB cannot be controlled by increasing population size and/or replication number.
Next steps: Develop and test approaches to control the impacts of GDMB.
Yan Zhou, Aaron Kusmec, Seyed Vahid Mirnezami, Lakshmi Attigala, Srikant Srinivasan, Talukder Zaki Jubery, James C. Schnable, Maria G. Salas Fernandez, Baskar Ganapathysubramanian, Patrick S. Schnable (2021). Identification and utilization of genetic determinants of trait measurement errors in image-based, high-throughput phenotyping. Plant Cell https://doi.org/10.1093/plcell/koab134