A Toolkit for Proximity-Dependent Biotinylation in Plants
In a systematic multi-lab study, Arora et al. apply a variety of proximity-dependent biotin labelling approaches using several different plant systems and bait proteins, to demonstrate its wide applicability for the plant community. Plant Cell https://doi.org/10.1105/tpc.20.00235.
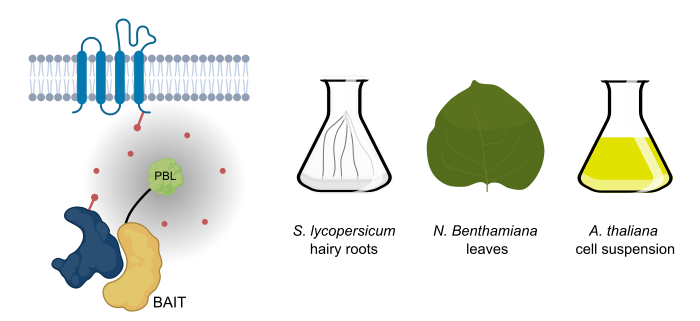
Background: Protein-protein interaction studies rely on preserving the interaction between the bait and prey proteins throughout the process of cell lysis, protein extraction, and protein complex purification. Preservation of the interaction and identifying the prey proteins is difficult in cases of low-affinity interactions, and particularly for integral membrane proteins as solubilizing them involves harsh conditions. Proximity labelling relies on in-vivo covalent tagging of prey proteins present in the near vicinity of the bait. Therefore, the interaction between the bait and prey need not be preserved during the process of protein purification. Finally, streptavidin-biotin pairing allows for selectively isolating the interactors and enables the identification of low-affinity interactions.
Question: As this tool was discovered and is extensively studied in animals, we aimed to establish proximity labelling across different plant model systems, such as Arabidopsis, Nicotiana benthamiana, and Solanum lycopersicum.
Findings: We found that out of the three generations of proximity tags available to date, the third generation, i.e., TurboID show more robustness in labelling interactors with exogenous biotin supplementation. Using NRF5 as a bait protein, we showed that this tool could be successfully used to capture the plasma membrane interactome of integral membrane proteins in plants. We identified and confirmed new interactors of the multi-subunit TPLATE complex, showing that this tool can expand our knowledge of interactomes beyond the currently used Affinity Purification-Mass Spectrometry-based approaches. Our protocol also enables us to find biotinylated peptides as direct proof of biotinylation. We provide the community with an extensive toolkit to use proximity labeling as an interactomics tool in different plant model systems, which provides the possibility to detect transient protein complex constituents.
Next steps: This tool is independent of protein solubility or protein complexation, hence applicable for interactomics studies of membrane proteins and intracellular compartments, providing a major advantage over alternative approaches. Distribution of identified biotinylated peptides, as well as their absence, will be significant in providing topology information in cases of transmembrane proteins and will aid in obtaining structural insight into protein complexes.
Deepanksha Arora, Nikolaj B. Abel, Chen Liu, Petra Van Damme, Klaas Yperman, Lam Dai Vu, Jie Wang, Anna Tornkvist, Francis Impens, Barbara Korbei, Dominique Eeckhout, Jelle Van Leene, Alain Goossens, Geert De Jaeger, Thomas Ott, Panagiotis Moschou, and Daniël Van Damme. (2020). Establishment of proximity-dependent biotinylation approaches in different plant model systems. Plant Cell. https://doi.org/10.1105/tpc.20.00235