Plant Science Research Weekly: September 25, 2020
Review: The hornworts: morphology, evolution and development
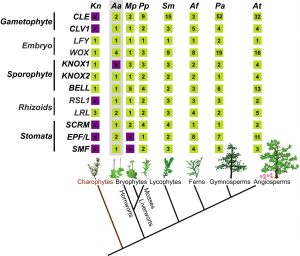 Bryophytes and vascular plants share a land plant as a common ancestor, but they have evolved independently for more than 400 million years. Recent genomic studies of model bryophytes, particularly mosses and liverworts, have provided insights into this ancient common ancestor. Here, Frangedakis et al. profile the newest model bryophyte, the hornwort Anthoceros agrestis, which harbors a number of unusual features for a land plant, including a continuously-growing sporangium, usually a single chloroplast per cell that may include a pyrenoid, and symbiotic relationships with endophytic cyanobacteria as well as mycorrhizal fungi. This review discusses each of these features as well as the hornwort lifecycle and development, and what is known about each of their molecular bases as compared to other model species. (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.16874
Bryophytes and vascular plants share a land plant as a common ancestor, but they have evolved independently for more than 400 million years. Recent genomic studies of model bryophytes, particularly mosses and liverworts, have provided insights into this ancient common ancestor. Here, Frangedakis et al. profile the newest model bryophyte, the hornwort Anthoceros agrestis, which harbors a number of unusual features for a land plant, including a continuously-growing sporangium, usually a single chloroplast per cell that may include a pyrenoid, and symbiotic relationships with endophytic cyanobacteria as well as mycorrhizal fungi. This review discusses each of these features as well as the hornwort lifecycle and development, and what is known about each of their molecular bases as compared to other model species. (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.16874
Review: La vie en roses, lilies, and other flowers: the floral microbiome
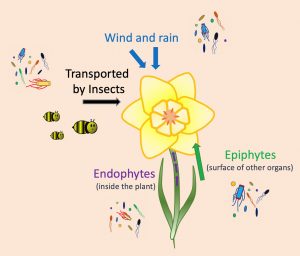 Beyond their beautiful colors, fragrances and shapes, flowers also can host a great variety of life forms. A diverse community of bacteria, fungi, protozoa, and other organisms live in the flowers. A recent work by Rachel Vannette summarizes the current studies about the floral microbiome, highlighting its importance at the ecological and evolutionary level. The first step to establish a microbial community is to arrive; microorganisms use the wind, rain, and insects as vehicles to reach the flowers. Some microbes live on leaves, shoot, and other organs of the plant (epiphytes), so can colonize the flowers as soon as they bloom. Other species live inside (endophytes) and can reach the flowers by traveling through the vascular tissues. The next step is to survive. The composition of the microbiome is very dynamic, with selection and competition among species two of the causes of this variation. Additionally, some flower traits such as the release of chemicals or morphological features can affect microbial growth. The flower microbiome can alter the quantity and quality of the nectar, or produce volatile compounds that, in the end, may affect pollination. This work summarizes progress on the characterization of the flower microbiomes. It describes the commonly used techniques to study it: in vitro culture, microscopy, and sequencing. Finally, the author presents some questions driving current research on this topic, for instance, “to what extent do microbes drive the evolution of floral traits?” (Summary by Humberto Herrera-Ubaldo @herrera_h) Annu. Rev. Ecol. Evol. Syst. 10.1146/annurev-ecolsys-011720-013401
Beyond their beautiful colors, fragrances and shapes, flowers also can host a great variety of life forms. A diverse community of bacteria, fungi, protozoa, and other organisms live in the flowers. A recent work by Rachel Vannette summarizes the current studies about the floral microbiome, highlighting its importance at the ecological and evolutionary level. The first step to establish a microbial community is to arrive; microorganisms use the wind, rain, and insects as vehicles to reach the flowers. Some microbes live on leaves, shoot, and other organs of the plant (epiphytes), so can colonize the flowers as soon as they bloom. Other species live inside (endophytes) and can reach the flowers by traveling through the vascular tissues. The next step is to survive. The composition of the microbiome is very dynamic, with selection and competition among species two of the causes of this variation. Additionally, some flower traits such as the release of chemicals or morphological features can affect microbial growth. The flower microbiome can alter the quantity and quality of the nectar, or produce volatile compounds that, in the end, may affect pollination. This work summarizes progress on the characterization of the flower microbiomes. It describes the commonly used techniques to study it: in vitro culture, microscopy, and sequencing. Finally, the author presents some questions driving current research on this topic, for instance, “to what extent do microbes drive the evolution of floral traits?” (Summary by Humberto Herrera-Ubaldo @herrera_h) Annu. Rev. Ecol. Evol. Syst. 10.1146/annurev-ecolsys-011720-013401
Review: The pleurogram, an under-investigated functional trait in seeds
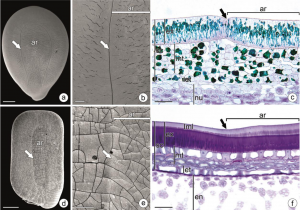 The pleurogram is a depression on both sides of the seeds of some Fabaceae species. Surprisingly, although it is recognized as a specialized structure, little is known about its anatomical features or biological role. In this review, Rodrigues-Junior et al. synthesize the current knowledge about the structure and function of the pleurogram. The pleurogram is present in at least 31 genera of Fabaceae, either as a single large depression covering most of the seeds’ surface or multiple tiny ones scattered over it. Moreover, they can arise either from a mechanic rupture of the testa’s palisade layer or a difference in this tissue’s cells’ height. Classical studies suggested that the pleurogram acts as a hygroscopic valve, meaning it opens and closes in response to environmental humidity. However, the authors note that the structure becomes fully closed when the seed reaches maturity and dormancy, and only opens again when dormancy-breaking conditions are met so that seeds can imbibe and germinate. Furthermore, some species possess a non-functional pleurogram, casting questions about the biological implications of gain/loss of its function. This review should spur efforts to increase understanding of this seed feature. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Ann. Bot. 10.1093/aob/mcaa161
The pleurogram is a depression on both sides of the seeds of some Fabaceae species. Surprisingly, although it is recognized as a specialized structure, little is known about its anatomical features or biological role. In this review, Rodrigues-Junior et al. synthesize the current knowledge about the structure and function of the pleurogram. The pleurogram is present in at least 31 genera of Fabaceae, either as a single large depression covering most of the seeds’ surface or multiple tiny ones scattered over it. Moreover, they can arise either from a mechanic rupture of the testa’s palisade layer or a difference in this tissue’s cells’ height. Classical studies suggested that the pleurogram acts as a hygroscopic valve, meaning it opens and closes in response to environmental humidity. However, the authors note that the structure becomes fully closed when the seed reaches maturity and dormancy, and only opens again when dormancy-breaking conditions are met so that seeds can imbibe and germinate. Furthermore, some species possess a non-functional pleurogram, casting questions about the biological implications of gain/loss of its function. This review should spur efforts to increase understanding of this seed feature. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Ann. Bot. 10.1093/aob/mcaa161
Opinion. We have been in lockdown, but deforestation has not
 Douglas Daly is the B. A. Krukoff Curator of Amazonian Botany, Institute of Systematic Botany at the New York Botanic Garden, and a specialist in tropical tree systematics and the flora of the Amazon region. In this Opinion article, he reflects on how he and other field-based botanists have kept their research moving forward, depending on local experts and digitized specimens. Yet he finds that the ongoing deforestation has not slowed during the pandemic but has instead increased to the rate of “more than half a square mile of deforestation per hour.” Deforestation is of global as well as of regional concern: “Globally, the Amazon’s trees—still in the billions—absorb 2 billion tons of CO2 per year, or 5% of annual emissions, making it a vital buffer against climate change.” Expanded funding sources and training programs are needed to help ensure the long-term protection of these critical environments once the botanists are once again able to get their boots on the ground – and up on the trees. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2018489117
Douglas Daly is the B. A. Krukoff Curator of Amazonian Botany, Institute of Systematic Botany at the New York Botanic Garden, and a specialist in tropical tree systematics and the flora of the Amazon region. In this Opinion article, he reflects on how he and other field-based botanists have kept their research moving forward, depending on local experts and digitized specimens. Yet he finds that the ongoing deforestation has not slowed during the pandemic but has instead increased to the rate of “more than half a square mile of deforestation per hour.” Deforestation is of global as well as of regional concern: “Globally, the Amazon’s trees—still in the billions—absorb 2 billion tons of CO2 per year, or 5% of annual emissions, making it a vital buffer against climate change.” Expanded funding sources and training programs are needed to help ensure the long-term protection of these critical environments once the botanists are once again able to get their boots on the ground – and up on the trees. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2018489117
A cross-kingdom conserved ER-phagy receptor maintains ER homeostasis during stress
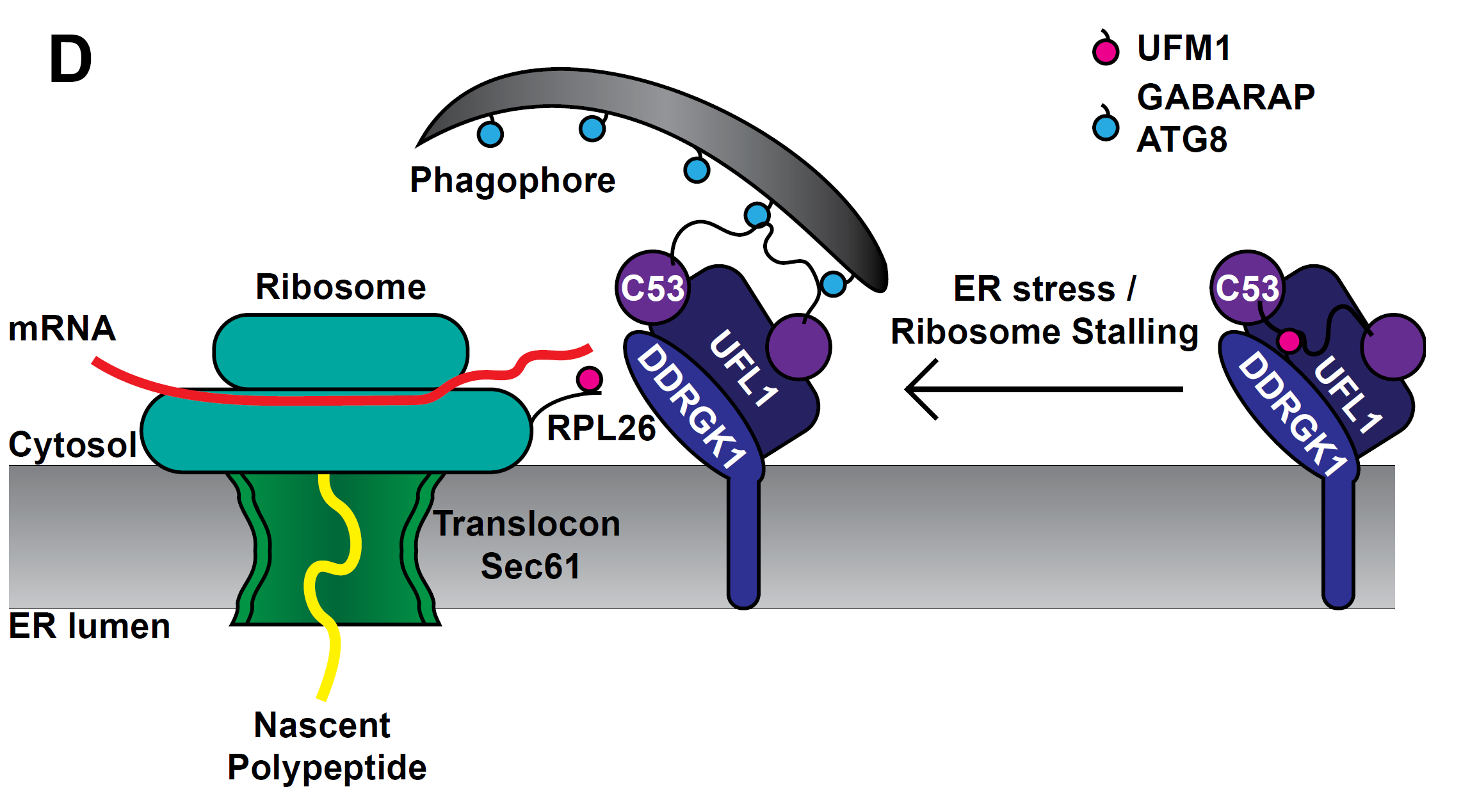 Quality control of the protein folding mechanism in the endoplasmic reticulum (ER), which selectively eliminates or recycles unwanted cytoplasmic components, is recognized by specific autophagy receptors. Selective removal of certain ER domains by the autophagy pathway (termed as ER-phagy) is controlled by these receptors that recruit specific cargo through interaction with the Autophagy-related protein 8 (ATG8) and then delivered for degradation and recycling. Stephani et al. identified a new receptor, C53, which is conserved across kingdoms that is specifically recruited into autophagosomes during ER stress. C53 binds ATG8 in A. thaliana, Marchantia polymorpha and human HeLa cells, and C53 localizes to the transport vesicles during ER stress. C53-ATG8 interaction is mediated via C53 conserved intrinsically disorder region with a novel consensus motif “IDWG” which is located at the highly flexible region. Interestingly, this C53 is neither activated by association with unfolded protein response sensors or ER-associated degradation pathway substrates, nor with the signal recognition particle. C53 is only activated upon ribosome stalling during co-translational protein translocation. The C53 mutant in plants is highly sensitive to ER stress caused by phosphate starvation. Overall, this study sheds light on a unique ER-recycling receptor that maintains cellular quality control. (Summary by Min May Wong @wongminmay) eLife 10.7554/elife.58396
Quality control of the protein folding mechanism in the endoplasmic reticulum (ER), which selectively eliminates or recycles unwanted cytoplasmic components, is recognized by specific autophagy receptors. Selective removal of certain ER domains by the autophagy pathway (termed as ER-phagy) is controlled by these receptors that recruit specific cargo through interaction with the Autophagy-related protein 8 (ATG8) and then delivered for degradation and recycling. Stephani et al. identified a new receptor, C53, which is conserved across kingdoms that is specifically recruited into autophagosomes during ER stress. C53 binds ATG8 in A. thaliana, Marchantia polymorpha and human HeLa cells, and C53 localizes to the transport vesicles during ER stress. C53-ATG8 interaction is mediated via C53 conserved intrinsically disorder region with a novel consensus motif “IDWG” which is located at the highly flexible region. Interestingly, this C53 is neither activated by association with unfolded protein response sensors or ER-associated degradation pathway substrates, nor with the signal recognition particle. C53 is only activated upon ribosome stalling during co-translational protein translocation. The C53 mutant in plants is highly sensitive to ER stress caused by phosphate starvation. Overall, this study sheds light on a unique ER-recycling receptor that maintains cellular quality control. (Summary by Min May Wong @wongminmay) eLife 10.7554/elife.58396
POME: Quantitative and dynamic cell polarity tracking pipeline
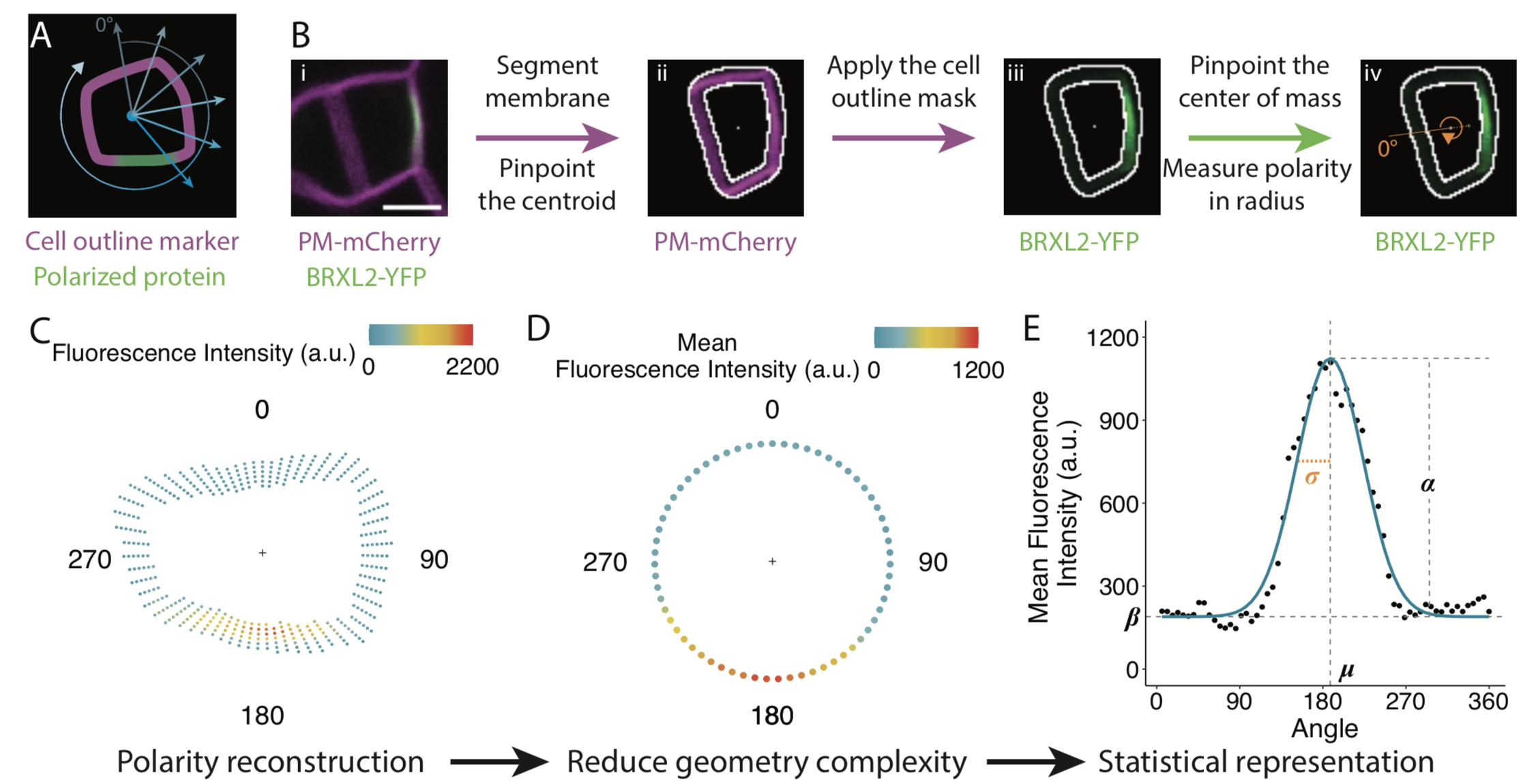 Many proteins polarize in the cell creating a subcellular niche for various functions. Asymmetric distribution of proteins is a general mechanism for localized growth, directional long-range signaling, cell migration, and asymmetric cell divisions. Well-known examples of polarity proteins include PIN-FORMED1 in auxin transport, and BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL), and BREVIS RADIX family (BRXf) in stomatal asymmetric cell divisions. Most data on polarized proteins are qualitative, without much information on protein quantity, size, and dynamics of the polar domain. In this paper, Gong et al. developed the POME (Polarity Mediated) pipeline for quantitative data analysis of polar proteins with good statistical power. POME is a semiautomated tool that requires a confocal image taken with two different channels (one must highlight the outline of the cell), the FIJI macro, and R script. The macro extracts the cell outline and determines both the amplitude and size of the polarity distribution. The authors have validated the POME using known polarized and depolarized proteins and show that BRXL2 accumulates prior to BASL in Arabidopsis. The authors also extended this analysis to other cell types (root cells) and model organisms (Brachypodium, C. elegans) to determine protein polarity. Thus, POME can be used in diverse systems to get quantitative information of polarity with statistical confidence. (Summary by Vijaya Batthula @Vijaya_Batthula) bioRxiv. 10.1101/2020.09.12.294942.
Many proteins polarize in the cell creating a subcellular niche for various functions. Asymmetric distribution of proteins is a general mechanism for localized growth, directional long-range signaling, cell migration, and asymmetric cell divisions. Well-known examples of polarity proteins include PIN-FORMED1 in auxin transport, and BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL), and BREVIS RADIX family (BRXf) in stomatal asymmetric cell divisions. Most data on polarized proteins are qualitative, without much information on protein quantity, size, and dynamics of the polar domain. In this paper, Gong et al. developed the POME (Polarity Mediated) pipeline for quantitative data analysis of polar proteins with good statistical power. POME is a semiautomated tool that requires a confocal image taken with two different channels (one must highlight the outline of the cell), the FIJI macro, and R script. The macro extracts the cell outline and determines both the amplitude and size of the polarity distribution. The authors have validated the POME using known polarized and depolarized proteins and show that BRXL2 accumulates prior to BASL in Arabidopsis. The authors also extended this analysis to other cell types (root cells) and model organisms (Brachypodium, C. elegans) to determine protein polarity. Thus, POME can be used in diverse systems to get quantitative information of polarity with statistical confidence. (Summary by Vijaya Batthula @Vijaya_Batthula) bioRxiv. 10.1101/2020.09.12.294942.
A peptide pair coordinates regular ovule initiation patterns with seed number and fruit size
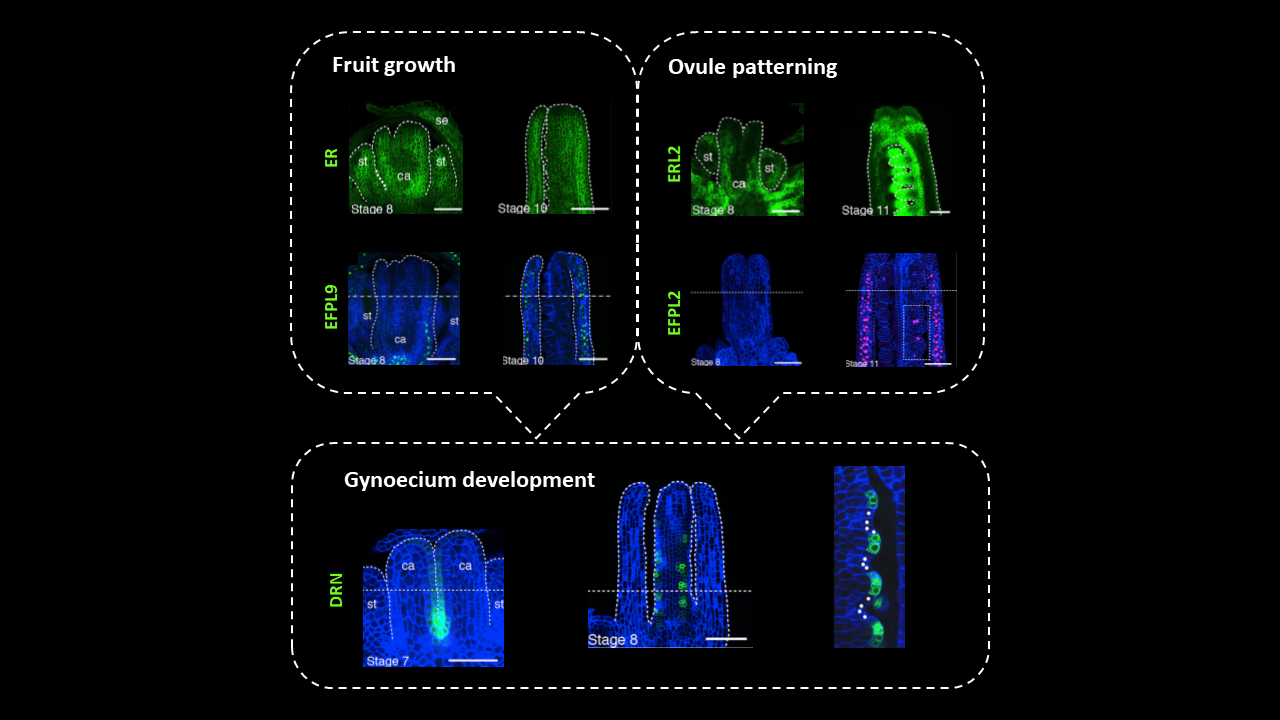
In Arabidopsis pistils, ovule primordia differentiate from the placenta – meristematic tissue within the two fused carpels – in parallel rows with a regular spacing of 2-4 cells. Since fruit growth and ovule formation determine the maximum number of seeds enclosed in a silique, these two processes should be tightly coordinated to ensure reproductive success. Yet the mechanism regulating pattern formation in the developing gynoecium is still unresolved. In this article, Kawamoto and coworkers investigated the genetic determinants of ovule initiation pattern by exploring natural variation in almost 100 Arabidopsis accessions. QTL analysis led to the identification of the factors controlling ovule and seed density: members of the ERECTA (ER) family of leucine-rich repeat receptor kinases, and their ligands belonging to EPIDERMAL PATTERNING FACTOR(EPF)/EPF-like (EPFL)-family of cysteine-rich secreted peptides. Genetic and expression analyses revealed roles for two separate signaling pathways. The first module relies on EPFL9 acting as a short-range signal from the carpel wall through ER and ER-like receptors to promote gynoecium growth without affecting the patterning of ovule initials. The second module relies on the interaction between EPFL2 and ERL1-ERL2 in the carpel wall and in inter-ovule spaces to control ovule density through equidistant spacing of ovule primordia. As supporting evidence, loss of EPFL2 function leads to irregular spacing and even ovule twinning. Thus, the differential expression of EPFL2 regulates spacing between adjacent ovules, which allows optimization of resource allocation and reduces competition among developing seeds after fertilization. (Summary and image adaptation by Michela Osnato @michela_osnato) Curr. Biol: 10.1016/j.cub.2020.08.050
Coordination of microbe-host homeostasis via a crosstalk with plant innate immunity
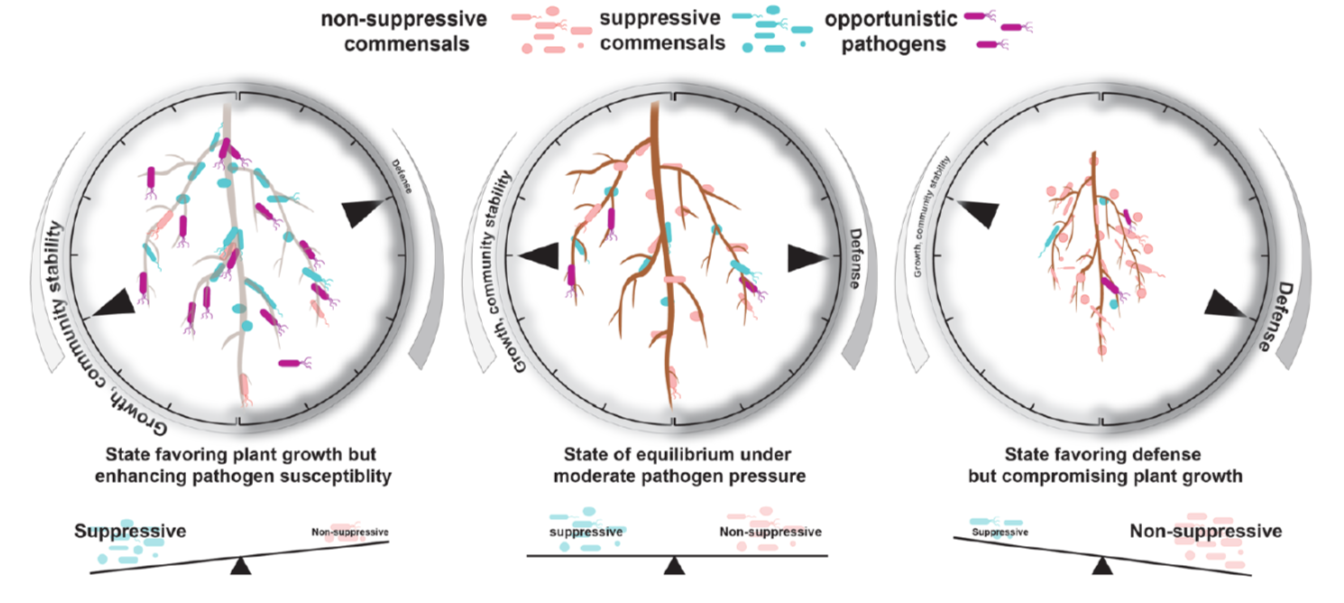
In nature, roots are colonized by commensal bacteria that do not seem to influence host traits. Recently, studies showed some commensals can suppress plant immune responses when inoculated to plants alone. However, how such commensal activities are coordinated in the community context is poorly understood. Ma, Niu et al. took a bottom-up approach to explore immune-modulating activities of various commensals and their interactions. The authors found that taxonomically diverse commensal strains (62 out of 151 strains) can interfere with root growth inhibition (RGI) induced by flg22 peptide, a microbe-derived immune elicitor. They then composed 5-member synthetic communities with suppressive or non-suppressive strains (suppressive/non-suppressive SynComs). They found that suppression of flg22-triggered RGI is, in some cases, associated with the ability of commensal strains to modify or degrade flg22 peptide. Flg22 treatment altered community structure of non-suppressive SynComs, but not suppressive SynComs, implying that flg22-induced immunity influences root microbiota establishment. Interestingly, mixtures of suppressive and non-suppressive SynComns was not affected by flg22 treatment, suggesting that RGI-suppression is the dominant trait in plant-associated bacterial communities and is thus easily overlooked in previous studies. Transcriptome analyses revealed a subset of known defense-related genes upregulated by a non-suppressive SynCom but downregulated by a suppressive SynCom. Furthermore, pre-colonization by non-suppressive SynComs protected plants from opportunistic pathogens, whereas suppressive SynComs did not. Together, the authors propose a model in which a balance between commensals with contrasting immune-modulating activities buffers plant resistance against pathogens and defense-associated plant growth reduction, eventually leading to microbe-host homeostasis. (Summary by Tatsuya Nobori @nobolly) Research Square 10.21203/rs.3.rs-69445/v1