A rice transcription factor controls grain length through cell number
Providing around 20% of global calories, rice (Oryza sativa) is one of the most important staple crops (Global Rice Science Partnership (GRiSP), 2013). With consumption particularly increasing in sub-Saharan Africa, the Middle East and Latin America, it is vital to increase rice yields to supply the growing population. Grain yield is largely determined by three characteristics; grain weight (of which grain size is a major contributing factor), number of grains per panicle, and number of panicles per plant (Li et al., 2018).
The identification of novel genes involved in grain yield enriches our understanding of the regulatory mechanisms behind this process and may ultimately contribute to the breeding of rice varieties with increased yield. Furthermore, a preference for longer grain length, including in several regions in China and India, could make the production of rice with increased grain length economically advantageous (Calingacion et al., 2014). Several quantitative trait loci (QTLs) contributing to grain size in rice have been identified, and act through a variety of mechanisms including phytohormone signalling, proteasomal degradation and transcription factors affecting downstream gene expression (reviewed in (Zuo and Li, 2014)). However, it remains unclear how these genes interact through their respective regulatory pathways, and what cross-talk exists between the pathways or within the network. It is hoped that the characterisation of further novel QTLs and investigation into their modes of action will help to uncover the molecular mechanisms controlling grain yield in rice.
 In this issue of Plant Physiology, Bin Han and colleagues from the Chinese Academy of Science and Shanghai Normal University present their characterisation of a transcription factor involved in grain size determination (Wang et al., 2019). During their previous work to illuminate the genetic origins of cultivated rice, the authors from the Chinese Academy of Science had discovered a major QTL (referred to as GL6) that underlies approximately 20% of the difference in grain length between the cultivated rice variety O. sativa ssp. indica Guangluai4 (GLA4) and the wild rice Oryza rufipogon W1943 (W1943; Huang et al., 2012). In the current publication, the researchers mapped the GL6 QTL to a gene encoding a plant AT-rich sequence and zinc- binding (PLATZ) transcription factor (GL6) using a GLA4 x W1943 line backcrossed to GLA4 (backcross inbred line BIL219). The GLA4 parent has longer rice grains and a full-length GL6 gene, while the W1943 parent is heterozygous for a premature stop codon close to the end of the region encoding the PLATZ domain. Several genetic approaches were used to verify the function of the gene. Lines with reduced GL6 function included a near isogenic line produced through further backcrossing to GLA4, which is homozygous for the premature stop codon (NIL-gl6), as well as CRISPR-Cas9-generated loss-of-function mutants. Complementation lines were also produced, in which the complete GL6 genetic region from GLA4 was introduced into NIL-gl6 via Agrobacterium-mediated transformation.
In this issue of Plant Physiology, Bin Han and colleagues from the Chinese Academy of Science and Shanghai Normal University present their characterisation of a transcription factor involved in grain size determination (Wang et al., 2019). During their previous work to illuminate the genetic origins of cultivated rice, the authors from the Chinese Academy of Science had discovered a major QTL (referred to as GL6) that underlies approximately 20% of the difference in grain length between the cultivated rice variety O. sativa ssp. indica Guangluai4 (GLA4) and the wild rice Oryza rufipogon W1943 (W1943; Huang et al., 2012). In the current publication, the researchers mapped the GL6 QTL to a gene encoding a plant AT-rich sequence and zinc- binding (PLATZ) transcription factor (GL6) using a GLA4 x W1943 line backcrossed to GLA4 (backcross inbred line BIL219). The GLA4 parent has longer rice grains and a full-length GL6 gene, while the W1943 parent is heterozygous for a premature stop codon close to the end of the region encoding the PLATZ domain. Several genetic approaches were used to verify the function of the gene. Lines with reduced GL6 function included a near isogenic line produced through further backcrossing to GLA4, which is homozygous for the premature stop codon (NIL-gl6), as well as CRISPR-Cas9-generated loss-of-function mutants. Complementation lines were also produced, in which the complete GL6 genetic region from GLA4 was introduced into NIL-gl6 via Agrobacterium-mediated transformation.
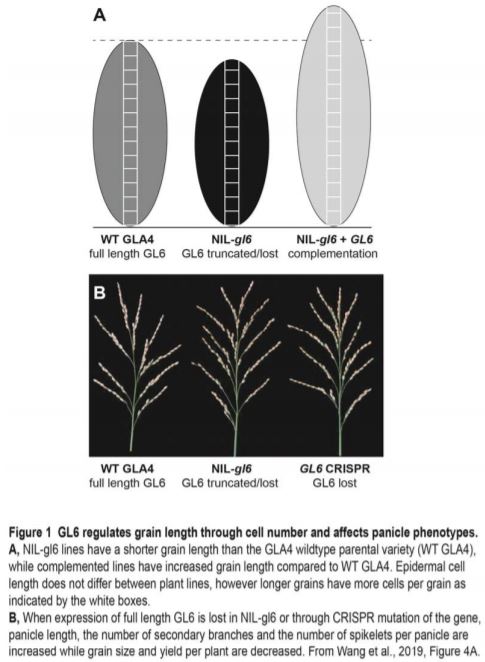
Taken together, the developmental analysis of these lines shows that GL6 positively regulates grain length and weight (summarised in Figure 1). Wang and colleagues demonstrated that grain length, and subsequently 1000-grain weight, were significantly decreased when expression of full-length GL6 was lost in NIL-gl6. By contrast, complementation of NIL-gl6 with full-length GL6 from GLA4 rescued the phenotype, with lines producing even longer grains than in the wild-type GLA4 parental variety. Increased length can be due to increased cell divisions resulting in more cells of a similar size, or increased cell elongation resulting in a similar number of longer cells, or a combination of the two. The authors found that cell length in the outer glume was not significantly different between the lines tested; however, the complementation line had a higher total cell number than the GLA4 wild type parental variety, which in turn had higher total cell number than mutant lines lacking a full-length GL6 gene (Figure 1A).
In an effort to explain why the cell number is increased in lines with a full-length GL6 gene, the expression levels of cell cycle genes were compared between GLA4 and NIL-gl6. Twelve of the 26 genes studied had significantly lower expression in NIL-gl6 compared to GLA4. The authors suggest that GL6 increases grain length through the alteration of cell division; however it is unclear whether these changes in gene expression reflect or are causative of the fewer cell divisions taking place when GL6 is truncated. Interestingly, GL6 expression in the complementation line is approximately four times higher than in the GLA4, and not significantly different between the wild-type GLA4 and NIL-gl6, although it must be noted that the transcript levels will not necessarily directly correlate with active protein levels, especially in the case of truncated transcripts. There is limited evidence that PLATZ transcription factors may have a role in the promotion of cell division in Arabidopsis thaliana leaves through interaction with the GROWTH REGULATING FACTOR (GRF) / GRF-INTERACTING FACTOR (GIF) regulatory module (Kim et al., 2018). This module regulates cell growth and proliferation, and is particularly important in the generation of stem cells for organogenesis (Kim and Tsukaya, 2015). Understanding whether this reflects a conserved mechanism in the angiosperms could potentially provide further causal evidence that links GL6 to cell number and hence grain length.
As discussed earlier, yield is dependent not only on grain weight, but also on grains per panicle and panicles per plant. A trade-off between grain weight and grain number has been noted for many plant species (Sadras, 2007), with involvement of a mitogen-activated protein kinase cascade linked to coordination of grain number and weight in rice (Guo et al., 2018). While GL6 positively regulates grain weight, increased expression also results in fewer secondary branches and fewer grains per panicle (Figure 1B). Tiller number increased only in the complementation line, where GL6 expression is higher than in the GLA4 variety. Given the significantly higher expression of GL6 in this line, it would be interesting to investigate if there is a causal link here. Furthermore, GL6 may reduce inflorescence meristem activity, as several genes that are known to delay the transition from inflorescence to spikelet meristem (Yoshida et al., 2013) were significantly upregulated in NIL-gl6. Overall, lines with either a reduction or overexpression of GL6 had lower yield than wild-type plants, indicating that GL6 could be an interesting target to alter grain length and thus satisfy consumer preferences for longer grains only if the yield penalty can be avoided.
When breeding rice plants for improved grain yield, it is vital that we increase our understanding of the trade-off between grain size and grain number. Yield is a complex trait that is unlikely to be improved through single gene changes without affecting other important agronomic traits. Although many QTLs underlying grain yield have been identified (Bai et al., 2012), and it has been demonstrated that biomass can be improved through QTL-based selection (e.g., Che et al., 2015), most QTLs are of relatively small effect when considered alone. Fortunately, there is an abundance of genetic material for crop improvement via QTL marker-assisted breeding available from several major rice germplasm banks throughout the world, including the International Rice Research Institute (IRRI), AfricaRice, the Centre for Tropical Agriculture (CIAT) and the Chinese Academy of Agricultural Sciences (CAAS). To best utilise these resources, a detailed understanding of the downstream effects of altering specific pathways and genes will be required to synergistically alter multiple points in the network. Interestingly, Wang and colleagues suggest that GL6 may interact with components of the RNA polymerase III complex, which could have broad downstream consequences for transcription and translation. As more genetic resources and techniques become available for rice, it would be interesting to further test these hypotheses in order to build a more robust understanding of pathways that determine grain length and yield in rice.
Naomi Cox
Department of Animal and Plant Sciences and The Plant Production and Protection (P3) Centre, University of Sheffield
ORCID ID: 0000-0002-0324-7995
Lisa M. Smith
Author for Contact
Department of Animal and Plant Sciences and The Plant Production and Protection (P3) Centre, University of Sheffield
ORCID ID: 0000-0003-2364-8187
References
Bai, X., Wu, B., and Xing, Y. (2012). Yield-related QTLs and their applications in rice. J. Integr. Plant Biol. 54, 300–311.
Calingacion, M., Laborte, A., Nelson, A., Resurreccion, A., Concepcion, J.C., Daygon, V.D., Mumm, R., and Reinke, R. (2014). Diversity of global rice markets and the science required for consumer-targeted rice breeding. PLoS One 9, e85106.
Che, R., Tong, H., Shi, B., Liu, Y., Fang, S., Liu, D., Xiao, Y., Hu, B., Liu, L., Wang, H., et al. (2015). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2, 1–7.
Global Rice Science Partnership (GRiSP) (2013). Rice Almanac.
Guo, T., Chen, K., Dong, N., Shi, C., Ye, W., Gao, J., and Shan, J. (2018). GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30, 871–888.
Huang, X., Kurata, N., Wei, X., Wang, Z., Wang, A., Zhao, Q., Zhao, Y., Liu, K., Dong, G., Zhan, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501.
Kim, J.H., and Tsukaya, H. (2015). Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J. Exp. Bot. 66, 6093–6107.
Kim, J.H., Kim, J., Jun, S.E., Park, S., Timilsina, R., Kwon, D.S., Park, S., Hwang, J.Y., Nam, H.G., Kim, G., et al. (2018). ORESARA15 , a PLATZ transcription factor , mediates leaf growth and senescence in Arabidopsis. New Phytol. 220, 609–623.
Li, N., Xu, R., Duan, P., and Li, Y. (2018). Control of grain size in rice. Plant Reprod. 31, 237–251.
Sadras, V.O. (2007). Evolutionary aspects of the trade-off between seed size and number in crops. F. Crop. Res. 100, 125–138.
Wang, A., Hou, Q., Si, L., Huang, X., and Luo, J. (2019). GL6, a PLATZ transcription factor, controls grain length and number in rice. Plant Physiol.
Yoshida, A., Sasao, M., Yasuno, N., Takagi, K., Daimon, Y., and Chen, R. (2013). TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. 110, 767–772.
Zuo, J., and Li, J. (2014). Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48, 99–118.