A New Tool for Visualizing Protein-Protein Interactions
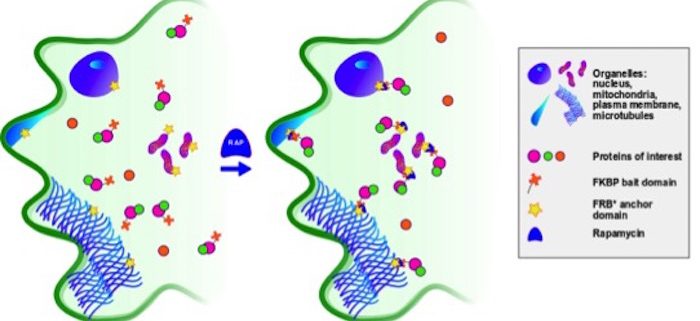
Background: Interactions between proteins (protein-protein interactions, PPIs) are an essential aspect of how life works. We aimed to design a tool with unique features and broad applications to test PPIs. Optimally, our tool would be free from some of the drawbacks of the other methods and would fill the gap in the field of PPIs that can be visualized using fluorescence microscopy. The tool needed to be suitable for proteins of different biological functions, and PPI visualization needed to be possible in different compartments of the cell. We wanted it to work quickly and conditionally, meaning that PPI visualization can be triggered on demand. Finally, we wanted to be able to quantify the observed interactions easily.
Question: Can we design a conditional protein-protein interaction tool that will meet all our criteria and work transiently in Nicotiana benthamiana epidermal leaf cells–a standard and commonly used plant model system?
Findings: We based our work on a pre-existing method in the animal field named Knocksideways (KS). In KS, the drug rapamycin causes the dimerization of two protein domains and allows proteins and their interaction partners to be sequestered. We implemented KS in plants (Knocksideways in plants, KSP), which meets all the criteria we aimed for. It is conditional and flexible. Visualization of PPIs can be triggered within minutes, and the PPIs remain visible for a prolonged period. KSP is suitable for visualizing diverse proteins with different functions and intracellular localizations. It allows the necessity of the interplay between multiple proteins for PPIs to be tested. KSP also has uses beyond a mere PPI assay, as it functions as a physiological assessment system. Finally, we designed custom-made scripts that allow the observations to be quantified. The availability of these scripts significantly reduces the time needed to analyze the results and draw conclusions.
Next steps: We plan to introduce KSP into different model plant species, such as Arabidopsis thaliana. This tool has the potential to generate functional knockouts in these plants at the protein level by conditionally removing proteins from their site of action in a complemented mutant background.
Joanna Winkler, Evelien Mylle, Andreas De Meyer, Benjamin Pavie, Julie Merchie, Peter Grones, Daniël Van Damme (2021). Visualizing protein-protein interactions in plants by rapamycin-dependent delocalization. Plant Cell https://doi.org/10.1093/plcell/koab004