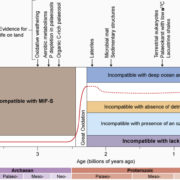
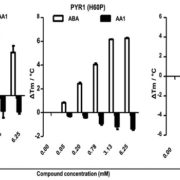
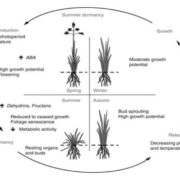
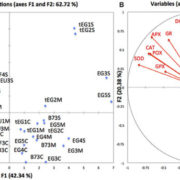
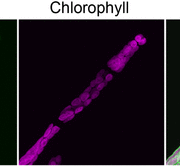
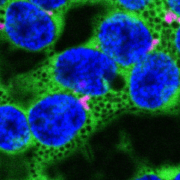
A dominant mutation in β-AMYLASE1 disrupts nighttime control of starch degradation in Arabidopsis leaves (Plant Physiol)
 Plants use starch in leaf chloroplasts to fuel their growth and metabolism during the night. In Arabidopsis, nighttime starch degradation is tightly regulated; the degradation rate is linear and adjusts to both the availability of starch and length of night, so that most but not all the starch is used up by dawn. The rate, S/T, is determined by a hypothetical “S” which tracks the amount of starch remaining, and “T”, the time till dawn sensed by the plant. Here, Feike et al. identified the bam1-2D mutant of BETA-AMYLASE1 (BAM1), which has a dominant missense mutation (S132N) causing accelerated starch degradation and premature exhaustion of starch reserves, possibly due to an uncoupling of S from the actual starch content. Mutant studies show that BAM1 transcript levels, protein content or phosphorylation state are not the cause, and in vitro enzymatic activity is not affected either. Instead, the strong phenotype could be caused by conformational changes leading to altered protein-protein interactions of a complex in vivo. Previous studies showed that BAM1 associates with a putative scaffold protein, LIKE SEX FOUR1 (LSF1), and that lsf1 mutants have a starch excess phenotype. Here, native gels, immunoblots and starch degradation assays from leaf extracts of various starch degradation mutants show an enhanced association of BAM1(S132N) with LSF1 compared to wild-type BAM1, and that this association is necessary for the accelerated starch degradation phenotype. Interestingly, bam1-2D lsf1 double mutants restore starch degradation to wild type rates, indicating that the bam1-2D mutation also overcomes the need for LSF1 in regular starch degradation. (Summary by Jiawen Chen @Jiaaawen) Plant Physiol. 10.1093/plphys/kiab603
Plants use starch in leaf chloroplasts to fuel their growth and metabolism during the night. In Arabidopsis, nighttime starch degradation is tightly regulated; the degradation rate is linear and adjusts to both the availability of starch and length of night, so that most but not all the starch is used up by dawn. The rate, S/T, is determined by a hypothetical “S” which tracks the amount of starch remaining, and “T”, the time till dawn sensed by the plant. Here, Feike et al. identified the bam1-2D mutant of BETA-AMYLASE1 (BAM1), which has a dominant missense mutation (S132N) causing accelerated starch degradation and premature exhaustion of starch reserves, possibly due to an uncoupling of S from the actual starch content. Mutant studies show that BAM1 transcript levels, protein content or phosphorylation state are not the cause, and in vitro enzymatic activity is not affected either. Instead, the strong phenotype could be caused by conformational changes leading to altered protein-protein interactions of a complex in vivo. Previous studies showed that BAM1 associates with a putative scaffold protein, LIKE SEX FOUR1 (LSF1), and that lsf1 mutants have a starch excess phenotype. Here, native gels, immunoblots and starch degradation assays from leaf extracts of various starch degradation mutants show an enhanced association of BAM1(S132N) with LSF1 compared to wild-type BAM1, and that this association is necessary for the accelerated starch degradation phenotype. Interestingly, bam1-2D lsf1 double mutants restore starch degradation to wild type rates, indicating that the bam1-2D mutation also overcomes the need for LSF1 in regular starch degradation. (Summary by Jiawen Chen @Jiaaawen) Plant Physiol. 10.1093/plphys/kiab603