A conserved ABA signaling module regulates dormancy in liverworts (Curr Biol)
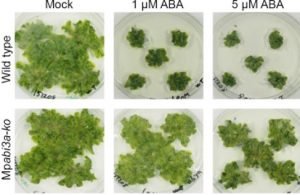 Plants can strategically maintain a state of dormancy to prevent germination and growth under unfavorable environmental conditions. In evolutionarily young lineages, dormancy is regulated by a canonical abscisic acid signalling pathway comprised of ABA receptors (PYR/PYL/RCAR), the negative regulator protein phosphatase 2C (PP2C) ABI1 (Abscisic acid Insensitive1), SNF1-related protein kinase2 (SnrK2), and ABA-associated transcriptional machinery (ABI3). Early-diverging land plant lineages like bryophytes (mosses, liverworts, and hornworts) also employ the ABA signalling module, although a functional role in regulating bryophyte dormancy is not well described. Eklund et al. reveal an evolutionarily conserved role for ABA in maintaining the dormancy of gemmae (clonal disk-like propagules housed in gemma cups) in the model liverwort Marchantia polymorpha. The authors demonstrate that exogenous ABA treatment exerts a dose-dependent suppressive effect on gemmaling development analogous to ABA-mediated dormancy in higher plants. Ectopic expression of the conserved MpABI1 regulatory protein led to ABA insensitivity and the release of gemmae dormancy. Moreover, ectopic expression of the conserved MpABI3 transcription factor activated conserved ABA-responsive loci in the absence of ABA, resulting in a stunted morphology that resembled that of wild-type liverworts exposed to high levels of ABA. Knockout mutants of Mpabi3 displayed enhanced tolerance to exogenous ABA and exhibited a loss of gemmae dormancy, which is consistent with the idea that ABI3 positively enforces ABA signalling in Marchantia. Collectively, these data demonstrate that the ABA signalling module is an evolutionarily conserved regulatory network that plays a key role in maintaining plant dormancy across a diverse range of tissue types present in distantly-related land plants. (Summary by Phil Carella) Curr. Biol. 10.1016/j.cub.2018.10.018
Plants can strategically maintain a state of dormancy to prevent germination and growth under unfavorable environmental conditions. In evolutionarily young lineages, dormancy is regulated by a canonical abscisic acid signalling pathway comprised of ABA receptors (PYR/PYL/RCAR), the negative regulator protein phosphatase 2C (PP2C) ABI1 (Abscisic acid Insensitive1), SNF1-related protein kinase2 (SnrK2), and ABA-associated transcriptional machinery (ABI3). Early-diverging land plant lineages like bryophytes (mosses, liverworts, and hornworts) also employ the ABA signalling module, although a functional role in regulating bryophyte dormancy is not well described. Eklund et al. reveal an evolutionarily conserved role for ABA in maintaining the dormancy of gemmae (clonal disk-like propagules housed in gemma cups) in the model liverwort Marchantia polymorpha. The authors demonstrate that exogenous ABA treatment exerts a dose-dependent suppressive effect on gemmaling development analogous to ABA-mediated dormancy in higher plants. Ectopic expression of the conserved MpABI1 regulatory protein led to ABA insensitivity and the release of gemmae dormancy. Moreover, ectopic expression of the conserved MpABI3 transcription factor activated conserved ABA-responsive loci in the absence of ABA, resulting in a stunted morphology that resembled that of wild-type liverworts exposed to high levels of ABA. Knockout mutants of Mpabi3 displayed enhanced tolerance to exogenous ABA and exhibited a loss of gemmae dormancy, which is consistent with the idea that ABI3 positively enforces ABA signalling in Marchantia. Collectively, these data demonstrate that the ABA signalling module is an evolutionarily conserved regulatory network that plays a key role in maintaining plant dormancy across a diverse range of tissue types present in distantly-related land plants. (Summary by Phil Carella) Curr. Biol. 10.1016/j.cub.2018.10.018