What We’re Reading: November 9th
Review: Single-particle tracking for the quantification of membrane protein dynamics in living plant cells
![]() Real-time tracking is a hugely powerful way to understand the behavior of single proteins. Cui et al. review the methods and applications of single-particle tracking (SPT) in plant cells. They describe the applications of variable-angle total internal reflection fluorescence microscopy (VA-TIRFM) to visualize fluorescently labeled plasma-membrane proteins. SPT has revealed many interesting and important aspects of membrane protein dynamics, such as changes in diffusion rate within the plasma membrane in response to activation or ligand binding. Protein oligomerization can also be detected through stepwise fluorescence bleaching, endocytosis can be observed (and dwell-time at the membrane calculated) by disappearance of fluorescent signals from the membrane, and co-localization dynamics between proteins observed. The authors conclude, “It is anticipated that in the near future these approaches will fundamentally change our views of the molecular processes actually occurring in living cells.” (Summary by Mary Williams) Mol. Plant 10.1016/j.molp.2018.09.008
Real-time tracking is a hugely powerful way to understand the behavior of single proteins. Cui et al. review the methods and applications of single-particle tracking (SPT) in plant cells. They describe the applications of variable-angle total internal reflection fluorescence microscopy (VA-TIRFM) to visualize fluorescently labeled plasma-membrane proteins. SPT has revealed many interesting and important aspects of membrane protein dynamics, such as changes in diffusion rate within the plasma membrane in response to activation or ligand binding. Protein oligomerization can also be detected through stepwise fluorescence bleaching, endocytosis can be observed (and dwell-time at the membrane calculated) by disappearance of fluorescent signals from the membrane, and co-localization dynamics between proteins observed. The authors conclude, “It is anticipated that in the near future these approaches will fundamentally change our views of the molecular processes actually occurring in living cells.” (Summary by Mary Williams) Mol. Plant 10.1016/j.molp.2018.09.008
Review. Functional phenomics: An emerging field integrating high-throughput phenotyping, physiology, and bioinformatics
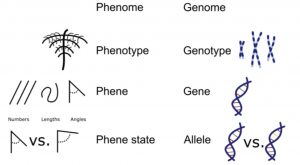 Understanding the processes contributing to plant productivity will help to secure food production in the future. Larry York proposes how this can be achieved using functional phenomics. First the ideotype needs to be constructed; the ideotype is a set of phene states that are hypothesized to enhance plant performance in a given environment. The ideotype determines the phenotyping platform to be used, which not only measures the phene state but also traits related to general plant performance such as plant biomass, and nutrient or water status. The selected phenes and indicators of overall plant performance can be subsequently studied in mapping populations, revealing relationships between phene and genetic loci. The multivariate approach helps to unlock the power of functional phenomics, revealing the relationship between the phene and function. Before the phene can be recommended as a target in breeding programs, it is necessary to functionally validate the contribution of the phene to plant performance by using, for example, molecular breeding techniques. (Summary by Magdalena Julkowska) J. Exp. Bot 10.1093/jxb/ery379
Understanding the processes contributing to plant productivity will help to secure food production in the future. Larry York proposes how this can be achieved using functional phenomics. First the ideotype needs to be constructed; the ideotype is a set of phene states that are hypothesized to enhance plant performance in a given environment. The ideotype determines the phenotyping platform to be used, which not only measures the phene state but also traits related to general plant performance such as plant biomass, and nutrient or water status. The selected phenes and indicators of overall plant performance can be subsequently studied in mapping populations, revealing relationships between phene and genetic loci. The multivariate approach helps to unlock the power of functional phenomics, revealing the relationship between the phene and function. Before the phene can be recommended as a target in breeding programs, it is necessary to functionally validate the contribution of the phene to plant performance by using, for example, molecular breeding techniques. (Summary by Magdalena Julkowska) J. Exp. Bot 10.1093/jxb/ery379
Heterohexamers formed by CcmK3 and CcmK4 increase the complexity of beta carboxysome shells
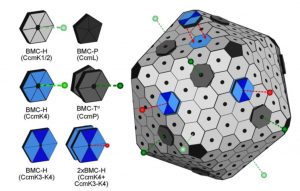 Carboxysomes are protein-bound microcompartments of cyanobacteria that sequester Rubisco and carbonic anhydrase (which converts bicarbonate to carbon dioxide), thus enhancing Rubisco’s carboxylation efficiency. The carboxysome shell is made up of hexamer, pentamer and trimer modules, with selectively permeable pores in the middle of each module. CcmK3 and CcmK4 are related proteins that contribute to the carboxysome shell, and there has been uncertainty as to whether these proteins act redundantly. Sommer et al. showed that CcmK4 can form a homohexamer or can form a heterohexamer with CcmK3, and that two heterohexamers can sometimes interact to form a “capped” structure. They authors propose that this capping may contribute to differential transport across the carboxysome shell and to the flexibility of carboxysomes under diverse conditions. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.01190
Carboxysomes are protein-bound microcompartments of cyanobacteria that sequester Rubisco and carbonic anhydrase (which converts bicarbonate to carbon dioxide), thus enhancing Rubisco’s carboxylation efficiency. The carboxysome shell is made up of hexamer, pentamer and trimer modules, with selectively permeable pores in the middle of each module. CcmK3 and CcmK4 are related proteins that contribute to the carboxysome shell, and there has been uncertainty as to whether these proteins act redundantly. Sommer et al. showed that CcmK4 can form a homohexamer or can form a heterohexamer with CcmK3, and that two heterohexamers can sometimes interact to form a “capped” structure. They authors propose that this capping may contribute to differential transport across the carboxysome shell and to the flexibility of carboxysomes under diverse conditions. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.01190
A conserved abscisic acid signaling module regulates dormancy in liverworts
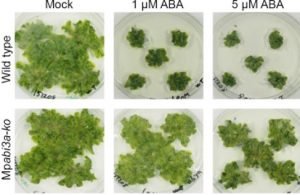 Plants can strategically maintain a state of dormancy to prevent germination and growth under unfavorable environmental conditions. In evolutionarily young lineages, dormancy is regulated by a canonical abscisic acid signalling pathway comprised of ABA receptors (PYR/PYL/RCAR), the negative regulator protein phosphatase 2C (PP2C) ABI1 (Abscisic acid Insensitive1), SNF1-related protein kinase2 (SnrK2), and ABA-associated transcriptional machinery (ABI3). Early-diverging land plant lineages like bryophytes (mosses, liverworts, and hornworts) also employ the ABA signalling module, although a functional role in regulating bryophyte dormancy is not well described. Eklund et al. reveal an evolutionarily conserved role for ABA in maintaining the dormancy of gemmae (clonal disk-like propagules housed in gemma cups) in the model liverwort Marchantia polymorpha. The authors demonstrate that exogenous ABA treatment exerts a dose-dependent suppressive effect on gemmaling development analogous to ABA-mediated dormancy in higher plants. Ectopic expression of the conserved MpABI1 regulatory protein led to ABA insensitivity and the release of gemmae dormancy. Moreover, ectopic expression of the conserved MpABI3 transcription factor activated conserved ABA-responsive loci in the absence of ABA, resulting in a stunted morphology that resembled that of wild-type liverworts exposed to high levels of ABA. Knockout mutants of Mpabi3 displayed enhanced tolerance to exogenous ABA and exhibited a loss of gemmae dormancy, which is consistent with the idea that ABI3 positively enforces ABA signalling in Marchantia. Collectively, these data demonstrate that the ABA signalling module is an evolutionarily conserved regulatory network that plays a key role in maintaining plant dormancy across a diverse range of tissue types present in distantly-related land plants. (Summary by Phil Carella) Curr. Biol. 10.1016/j.cub.2018.10.018
Plants can strategically maintain a state of dormancy to prevent germination and growth under unfavorable environmental conditions. In evolutionarily young lineages, dormancy is regulated by a canonical abscisic acid signalling pathway comprised of ABA receptors (PYR/PYL/RCAR), the negative regulator protein phosphatase 2C (PP2C) ABI1 (Abscisic acid Insensitive1), SNF1-related protein kinase2 (SnrK2), and ABA-associated transcriptional machinery (ABI3). Early-diverging land plant lineages like bryophytes (mosses, liverworts, and hornworts) also employ the ABA signalling module, although a functional role in regulating bryophyte dormancy is not well described. Eklund et al. reveal an evolutionarily conserved role for ABA in maintaining the dormancy of gemmae (clonal disk-like propagules housed in gemma cups) in the model liverwort Marchantia polymorpha. The authors demonstrate that exogenous ABA treatment exerts a dose-dependent suppressive effect on gemmaling development analogous to ABA-mediated dormancy in higher plants. Ectopic expression of the conserved MpABI1 regulatory protein led to ABA insensitivity and the release of gemmae dormancy. Moreover, ectopic expression of the conserved MpABI3 transcription factor activated conserved ABA-responsive loci in the absence of ABA, resulting in a stunted morphology that resembled that of wild-type liverworts exposed to high levels of ABA. Knockout mutants of Mpabi3 displayed enhanced tolerance to exogenous ABA and exhibited a loss of gemmae dormancy, which is consistent with the idea that ABI3 positively enforces ABA signalling in Marchantia. Collectively, these data demonstrate that the ABA signalling module is an evolutionarily conserved regulatory network that plays a key role in maintaining plant dormancy across a diverse range of tissue types present in distantly-related land plants. (Summary by Phil Carella) Curr. Biol. 10.1016/j.cub.2018.10.018
Major domestication-related phenotypes in indica rice are due to loss of miRNA-mediated laccase silencing
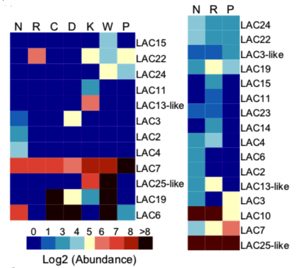 Rice (Oryza sativa) is derived from breeding of perennial wild ancestors with long stalks and few seeds to short plants with enlarged panicles (inflorescences). Many known domestication changes are due to changes in transcription factors or modulation of enzymatic actions. Recent work by Swetha et al. profiled small RNAs from multiple indica rice lines together with the wild species, and observed the unusual abundance primarily in the wild species of 22nt miRNA397, which results in selective degradation of laccases, which in turn contributes to reduction of stem lignification. Complementation with miRNA397 in cultivated rice lines shows a de-domestication phenotypic effect. miRNA397 is also known to be involved in increasing yield when overexpressed in japonica rice. Regulation of lignification by this miRNA also holds true for other crops. For example, S. pimpinellifolium, a wild relative of cultivated tomato, shows higher abundance of miR397 when compared to cultivated tomato lines. Hence this study sheds light on a previously unknown molecular phenomenon of miRNA regulated domestication-associated phenotypes in indica rice, and provides new tools for using miRNA397 as a probe in modern rice breeding programs. (Summary by Amey Redkar) Plant Cell: 10.1105/tpc.18.00472
Rice (Oryza sativa) is derived from breeding of perennial wild ancestors with long stalks and few seeds to short plants with enlarged panicles (inflorescences). Many known domestication changes are due to changes in transcription factors or modulation of enzymatic actions. Recent work by Swetha et al. profiled small RNAs from multiple indica rice lines together with the wild species, and observed the unusual abundance primarily in the wild species of 22nt miRNA397, which results in selective degradation of laccases, which in turn contributes to reduction of stem lignification. Complementation with miRNA397 in cultivated rice lines shows a de-domestication phenotypic effect. miRNA397 is also known to be involved in increasing yield when overexpressed in japonica rice. Regulation of lignification by this miRNA also holds true for other crops. For example, S. pimpinellifolium, a wild relative of cultivated tomato, shows higher abundance of miR397 when compared to cultivated tomato lines. Hence this study sheds light on a previously unknown molecular phenomenon of miRNA regulated domestication-associated phenotypes in indica rice, and provides new tools for using miRNA397 as a probe in modern rice breeding programs. (Summary by Amey Redkar) Plant Cell: 10.1105/tpc.18.00472
Contribution of epigenetic variation to adaptation in Arabidopsis
 It is known that changes in the epigenetic state can be inherited, but whether the epigenetic changes are subject to natural selection is uncharted territory. Schmid et al. examined the phenomic and epigenomic changes in Arabidopsis accessions that underwent selection to simulated habitat fragmentation. Five generations of selection resulted in alterations of plants’ phenotype and decreased epigenetic diversity among the selected lines. The low level of genome methylation corresponded to slower growth and later flowering than the ancestral lines. In particular, the methylation and expression of At2g06002 (encoding a non-coding RNA) corresponded to the late flowering phenotype. While the mechanisms underlying the environmentally induced changes in epigenome are unclear, this study shows that the selection can lead to changes in the epigenetic status and can play a role in stably inherited adaptive changes in phenotype. (Summary by Magdalena Julkowska) Nature Comms. 10.1038/s41467-018-06932-5
It is known that changes in the epigenetic state can be inherited, but whether the epigenetic changes are subject to natural selection is uncharted territory. Schmid et al. examined the phenomic and epigenomic changes in Arabidopsis accessions that underwent selection to simulated habitat fragmentation. Five generations of selection resulted in alterations of plants’ phenotype and decreased epigenetic diversity among the selected lines. The low level of genome methylation corresponded to slower growth and later flowering than the ancestral lines. In particular, the methylation and expression of At2g06002 (encoding a non-coding RNA) corresponded to the late flowering phenotype. While the mechanisms underlying the environmentally induced changes in epigenome are unclear, this study shows that the selection can lead to changes in the epigenetic status and can play a role in stably inherited adaptive changes in phenotype. (Summary by Magdalena Julkowska) Nature Comms. 10.1038/s41467-018-06932-5
Aphid-borne viral spread is enhanced by virus-induced accumulation of plant reactive oxygen species
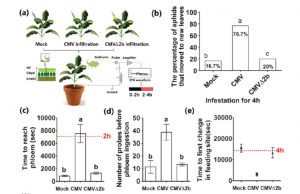 Aphids are major vectors for viral spread. Guo et al. have identified an interesting interaction that affects aphid feeding behavior and viral spread. They showed that reactive oxygen species (ROS) accumulation in plants infected with cucumber mosaic virus (CMV) affects aphid feeding behavior; specifically, it increases the frequency with which the stylets are inserted, delays penetration of phloem, and leads to increased aphid movement between leaves, all of which increase the exposure of the aphids to the virus. The contribution of ROS was demonstrated through exogenous ROS treatment of non-infected leaves. Furthermore, the viral suppressor protein VSR2b contributes to ROS production and therefore affects aphid feeding behavior. When plants are infected with CMV mutants lacking 2b (CMVΔ2b), ROS production is limited, aphids do not move as much, and viral spread is reduced. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00437
Aphids are major vectors for viral spread. Guo et al. have identified an interesting interaction that affects aphid feeding behavior and viral spread. They showed that reactive oxygen species (ROS) accumulation in plants infected with cucumber mosaic virus (CMV) affects aphid feeding behavior; specifically, it increases the frequency with which the stylets are inserted, delays penetration of phloem, and leads to increased aphid movement between leaves, all of which increase the exposure of the aphids to the virus. The contribution of ROS was demonstrated through exogenous ROS treatment of non-infected leaves. Furthermore, the viral suppressor protein VSR2b contributes to ROS production and therefore affects aphid feeding behavior. When plants are infected with CMV mutants lacking 2b (CMVΔ2b), ROS production is limited, aphids do not move as much, and viral spread is reduced. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00437
Microbial interkingdom interactions in roots promote Arabidopsis survival
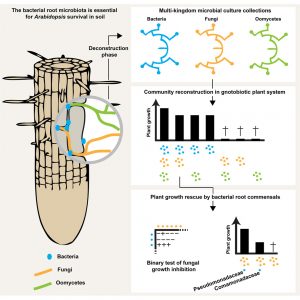 Plant roots in soil are surrounded by different microorganisms such as bacteria, fungi and oomycetes. The impact of these organisms on plant health has been known f0r many decades. However, the interaction network between these organisms is not well understood. By using next generation sequencing as a tool, Duran et al. profiled root associated multi-kingdom microbial communities in Arabidopsis. The data reveal that the relative abundance between bacterial and fungal community members are negatively correlated. Additionally, geographically distant Arabidopsis showed a similar bacterial microbial consortium while fungal and oomycete communities seemed to be distinct and site specific. This confirms plant specific effects on root-associated bacterial microbiota, which is consistent with previous reports. Selected bacterial, fungal and oomycete culture collection were used to reconstitute the microbiome under laboratory conditions, to investigate interkingdom competition. These studies suggest that plant root-associated bacterial populations execute a protection role to outcompete against the detrimental activities of filamentous eukaryotes, revealing their potential as biocontrol agents against filamentous pathogenic microbes. (Summary by Mugdha Sabale) Cell: 10.1016/j.cell.2018.10.020
Plant roots in soil are surrounded by different microorganisms such as bacteria, fungi and oomycetes. The impact of these organisms on plant health has been known f0r many decades. However, the interaction network between these organisms is not well understood. By using next generation sequencing as a tool, Duran et al. profiled root associated multi-kingdom microbial communities in Arabidopsis. The data reveal that the relative abundance between bacterial and fungal community members are negatively correlated. Additionally, geographically distant Arabidopsis showed a similar bacterial microbial consortium while fungal and oomycete communities seemed to be distinct and site specific. This confirms plant specific effects on root-associated bacterial microbiota, which is consistent with previous reports. Selected bacterial, fungal and oomycete culture collection were used to reconstitute the microbiome under laboratory conditions, to investigate interkingdom competition. These studies suggest that plant root-associated bacterial populations execute a protection role to outcompete against the detrimental activities of filamentous eukaryotes, revealing their potential as biocontrol agents against filamentous pathogenic microbes. (Summary by Mugdha Sabale) Cell: 10.1016/j.cell.2018.10.020



