When to Sleep? CHT7 is Critical for Nutrient-dependent Quiescence in Chlamydomonas
To survive nutrient scarcity, unicellular organisms establish a reversible quiescent state in which cells cease growth and division and switch metabolism to an energy-conserving mode. When nutrients become available again, cells exit from quiescence and resume growth and division. The mechanistic details for the establishment of an effective quiescent state in response to nutrient shortage remain poorly understood in photosynthetic eukaryotes. Takeuchi et al. (2020) studied the nutrient-dependent life cycle transition in a model green microalga, Chlamydomonas reinhardtii, to bridge this knowledge gap.
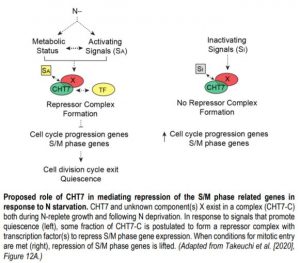 In Chlamydomonas, nitrogen (N) deprivation triggers the transition into quiescence accompanied by reprogramming of the transcriptome, proteome and metabolome (Schmollinger et al., 2014). A previous study by Tsai et al. (2014) identified a protein named COMPROMISED HYDROLYSIS OF TRIACYLGLYCEROLS 7 (CHT7) that affects the transcription of genes related to N deprivation-triggered quiescence. To gain further insight into the functional role of CHT7 in regulating the N-mediated life cycle transition of this alga, the authors re-evaluated the transcriptomic data of the cht7 mutant and its parental line (PL) previously collected by Tsai et al., (2014). While many genes involved in DNA metabolism and cell cycle regulation were repressed in PL cells following N deprivation, the expression of these genes, especially the S/M (DNA synthesis/mitosis) phase-specific genes, in N-deprived cht7 cells remained at a level similar to that in N-replete cht7 cells and slightly higher than that in N-replete PL cells. In addition, genes encoding nutrient transporters and oxidoreductases were downregulated in N-deprived cht7 when compared to PL during N starvation.
In Chlamydomonas, nitrogen (N) deprivation triggers the transition into quiescence accompanied by reprogramming of the transcriptome, proteome and metabolome (Schmollinger et al., 2014). A previous study by Tsai et al. (2014) identified a protein named COMPROMISED HYDROLYSIS OF TRIACYLGLYCEROLS 7 (CHT7) that affects the transcription of genes related to N deprivation-triggered quiescence. To gain further insight into the functional role of CHT7 in regulating the N-mediated life cycle transition of this alga, the authors re-evaluated the transcriptomic data of the cht7 mutant and its parental line (PL) previously collected by Tsai et al., (2014). While many genes involved in DNA metabolism and cell cycle regulation were repressed in PL cells following N deprivation, the expression of these genes, especially the S/M (DNA synthesis/mitosis) phase-specific genes, in N-deprived cht7 cells remained at a level similar to that in N-replete cht7 cells and slightly higher than that in N-replete PL cells. In addition, genes encoding nutrient transporters and oxidoreductases were downregulated in N-deprived cht7 when compared to PL during N starvation.
Does the misexpression of genes in cht7 cells result in phenotypic differences under various N conditions? Contrary to wild type and CHT7-complemented lines, cht7 cells failed to arrest growth and division and showed reduced cell viability during N deprivation. Following N resupply, cht7 cells showed delayed resumption of growth, abortive division, and a decline in the ability to form colonies. Therefore, the authors conclude that CHT7 plays an important role in the transcriptional establishment of N-responsive quiescence, which is critical for the viability of N-deprived cells and their resumption of growth and division following N refeeding. The authors noticed that the population of N-deprived cht7 cells prepared from a culture during mixotrophic growth was heterogeneous (e.g., some cells were metabolically active and some were not; some contained multiple nuclei per cell, while some had one nucleus). This raised an interesting question of whether the transcript levels in N-deprived cht7 cells represented a uniform expression across the total cell population or an average of varying expression levels in phenotypically different subpopulations. Future experiments such as transcriptional analyses of subpopulations, individual cells, or synchronized cultures of the cht7 mutant will be needed to address the interesting potential for variations amongst subpopulations in N-responsiveness.
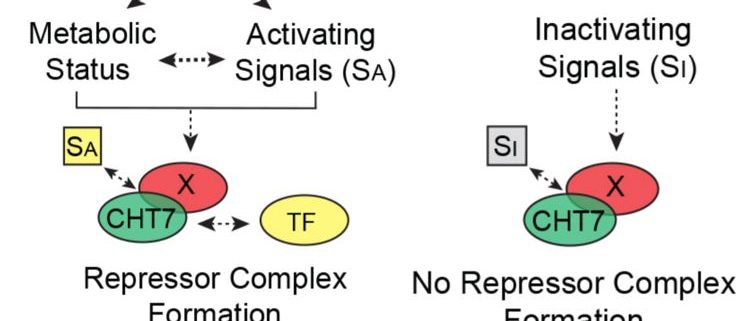
How does CHT7 mediate the transcriptional establishment of N-responsive quiescence? CHT7 contains two cysteine-rich (CXC) domains that have been shown in other organisms to be important for sequence-specific DNA binding. Intriguingly, removal of the CXC domains of CHT7 did not influence the ability of CHT7 to reverse the phenotypes and transcript levels of cht7 cells under different N conditions. On the other hand, the predicted protein-binding domains within the C-terminal half of CHT7 were found to be necessary for forming protein complexes, repressing cell cycle-related genes, and the proper entry into and exit from quiescence in response to N availability (see figure). These results suggest that CHT7 may serve as a scaffold for other transcriptional regulators, which is distinct from the role of conventional CXC domain-containing proteins in other organisms.
The identification of CHT7 as a key factor in the regulatory module responsible for cell cycle transitions paves the way for further explorations of the coordination between metabolic status and quiescence in green microalgae. Future studies on the heterogeneity of N-deprived cht7 cells as well as the composition of CHT7-mediated protein complexes will likely provide deeper insights into the molecular machinery for N-responsive quiescence.
Yingqi Cai
Biology Department, Brookhaven National Laboratory
ORCID: 0000-0002-0357-5809
REFERENCES
Schmollinger, S., Mühlhaus, T., Boyle, N.R., Blaby, I.K., Casero, D., Mettler, T., Moseley, J.L., Kropat, J., Sommer, F., Strenkert, D., Hemme, D., Pellegrini, M., Grossman, A.R., Stitt, M., Schroda, M., and Merchant, S.S. (2014). Nitrogen-Sparing Mechanisms in Chlamydomonas Affect the Transcriptome, the Proteome, and Photosynthetic Metabolism. Plant Cell 26: 1410-1435.
Takeuchi, T., Sears, B.B., Lindeboom, C., Lin, Y., Fekaris, N., Zienkiewicz, K., Zienkiewicz, A., Poliner, E., Benning, C. (2020) Chlamydomonas CHT7 is required for an effective quiescent state by regulating nutrient responsive cell cycle gene expression. Plant Cell, Published February 2020. DOI: https://doi.org/10.1105/tpc.19.00628.
Tsai, C.H., Warakanont, J., Takeuchi, T., Sears, B.B., Moellering, E.R., and Benning, C. (2014). The protein Compromised Hydrolysis of Triacylglycerols 7 (CHT7) acts as a repressor of cellular quiescence in Chlamydomonas. Proc. Natl. Acad. Sci. USA 111: 15833-15838.