When posttranslational modifications meet splicing to regulate stress responses
Agrofoglio et al. explore how the methylation of arginine residues on a key splicing factor affects alternative splicing.
Julieta Mateos, María José Iglesias, Yamila Agrofoglio. IFIBYNE-UBA-CONICET
https://doi.org/10.1093/plcell/koae051
Background: Plants are constantly subjected to a variety of environmental fluctuations, ranging from alterations in ambient temperature to infection by pathogens. In response to these stresses, plant cells trigger a complex reprogramming of gene expression, altering not only growth and development but also eliciting defense mechanisms. Several levels of gene regulation are involved in plant stress responses. These include transcriptional regulation, which determines which genes are activated; posttranscriptional regulation, which controls if mRNAs undergo alternative splicing; translational regulation, which controls when mRNAs are translated into proteins; and posttranslational regulation, which controls whether these proteins suffer additional modifications that influence their activity. Consequently, the interaction of the different molecular programs coordinating gene expression is essential for plant development.
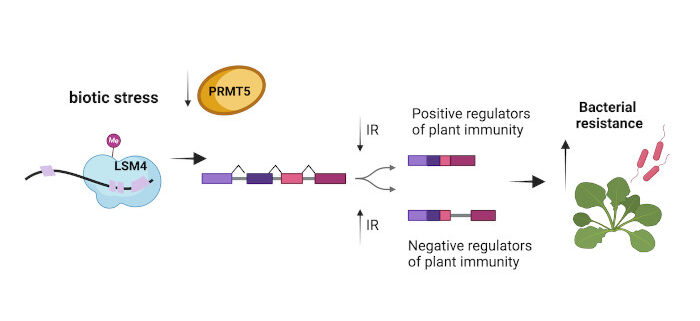
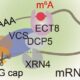
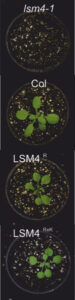 Question: Previous studies established that in Arabidopsis plants, the PRMT5 methyltransferase protein methylates the arginine residues of proteins that are involved in the regulation of alternative splicing. To determine the importance of methylation in alternative splicing regulation, especially in response to stress, we employed the SM-LIKE PROTEIN 4 (LSM4) splicing factor as a model.
Question: Previous studies established that in Arabidopsis plants, the PRMT5 methyltransferase protein methylates the arginine residues of proteins that are involved in the regulation of alternative splicing. To determine the importance of methylation in alternative splicing regulation, especially in response to stress, we employed the SM-LIKE PROTEIN 4 (LSM4) splicing factor as a model.
Findings: We show that loss of LSM4 leads to widespread missplicing, but its methylation has a restricted role in splicing under normal growth. On the other hand, LSM4 methylation responds to environmental cues associated with stress, showing antagonic response under both biotic and abiotic conditions. Bacterial infection leads to decreased LSM4 methylation, influencing the splicing of genes associated with plant immunity to enhance resistance. In contrast, salinity increases LSM4 methylation, crucial for the correct splicing of genes involved in abiotic stress responses.
Next steps: Our work shows how posttranslational modification changes RNA splicing to allow plants to acclimate to the environment. The interaction of diverse protein modifications, converging to efficiently control the RNA splicing process, has the potential to reveal key molecular mechanisms essential for plant adaptation.
Reference:
Yamila Carla Agrofoglio; María José Iglesias; Soledad Perez-Santángelo; María José de Leone; Tino Koester, Rafael Catalá; Julio Salinas; Marcelo J. Yanovsky; Dorothee Staiger; Julieta L. Mateos (2024) Arginine methylation of SM-LIKE PROTEIN 4 antagonistically affects alternative splicing during Arabidopsis stress responses. https://doi.org/10.1093/plcell/koae051