What We’re Reading: November 17th
J. Exp. Bot. Special Issue. The plant cuticle: old challenges, new perspectives ($)
 The cuticle is a cell-wall polymer that protects against desiccation, pathogens and UV light. Domínguez et al. provide an open-access editorial that describes this fine collection of articles covering all aspects of the plant cuticle, from its evolutionary origins to its ecological significance. Within the issue, article topics include the role of the cuticle in land plant colonization, cuticle’s physico-chemical and mechanical properties, and its contribution to organ separation and development, as well as the application of –omics tools to its study, and its potential in the production of bioplastics. (Summary by Mary Williams) https://academic.oup.com/jxb/issue/68/19
The cuticle is a cell-wall polymer that protects against desiccation, pathogens and UV light. Domínguez et al. provide an open-access editorial that describes this fine collection of articles covering all aspects of the plant cuticle, from its evolutionary origins to its ecological significance. Within the issue, article topics include the role of the cuticle in land plant colonization, cuticle’s physico-chemical and mechanical properties, and its contribution to organ separation and development, as well as the application of –omics tools to its study, and its potential in the production of bioplastics. (Summary by Mary Williams) https://academic.oup.com/jxb/issue/68/19
Review: Plant phenomics, from sensors to knowledge ($)
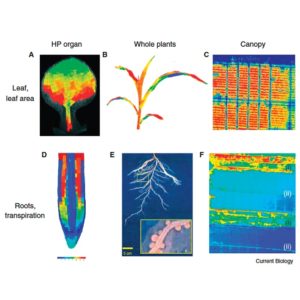
Plants adapt their form, function and metabolism to the surrounding environment. According to Tardieu et al., understanding the plant phenome requires that plant phenotypes to be studied on different spatial and temporal scales. High precision platforms are instrumental for discovery of new physiological mechanisms, but larger scale experiments are necessary to understand relationship to yield and interaction with environment. Once enough data is collected, the results can be used to develop models predicting plant phenotypes, generating new hypotheses, and revealing gaps in our understanding of how the plants work. In order to extend plant phenomics into the brave new world of big-data analysis, standardization of the protocols, data normalization and information provided alongside the experimental output is necessary. (Summary by Magdalena Julkowska) Curr. Biol. 10.1016/j.cub.2017.05.055
Review. Feed your friends: Do plant exudates shape the root microbiome? ($)
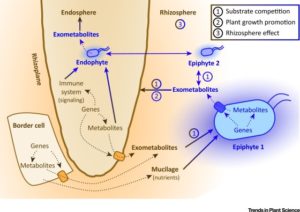 A wide variety of beneficial and pathogenic microbes surrounds plants both below and above ground. This microbial diversity is shaped by different biotic and abiotic factors and plays a role in maintaining plant health in natural settings. Hence, it is vital to understand the influence of different factors on determining the formulation of the microbial communities that associate with plants. Many studies have contributed to understanding the composition of rhizobiomes (microbes associated with roots), but fewer explore how plants shape their rhizobiomes. Sasse et al. review the contributions of plant genetic factors, root morphology and root exudation in shaping the rhizobiome, and explore a putative metabolite network between plant and microbes. They also summarize the transporters involved in root exudation and their contributions to the shaping of the rhizobiome. This article concludes by summarizing limitations to our current models, and outstanding questions including the need to analyze root exudation in natural settings. (Summary by Mugdha Sabale) Trends Plant Sci. 10.1016/j.tplants.2017.09.003
A wide variety of beneficial and pathogenic microbes surrounds plants both below and above ground. This microbial diversity is shaped by different biotic and abiotic factors and plays a role in maintaining plant health in natural settings. Hence, it is vital to understand the influence of different factors on determining the formulation of the microbial communities that associate with plants. Many studies have contributed to understanding the composition of rhizobiomes (microbes associated with roots), but fewer explore how plants shape their rhizobiomes. Sasse et al. review the contributions of plant genetic factors, root morphology and root exudation in shaping the rhizobiome, and explore a putative metabolite network between plant and microbes. They also summarize the transporters involved in root exudation and their contributions to the shaping of the rhizobiome. This article concludes by summarizing limitations to our current models, and outstanding questions including the need to analyze root exudation in natural settings. (Summary by Mugdha Sabale) Trends Plant Sci. 10.1016/j.tplants.2017.09.003
EIN3 and PIF3 interdependently repress chloroplast development in buried seedlings
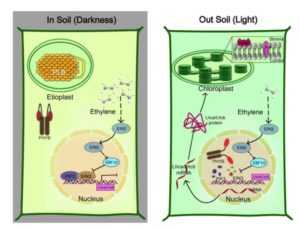 During embryogenesis, plastids arrest their differentiation as etioplasts (characterized by the distinctive prolamellar bodies, PLBs), and remain poised to complete their differentiation into functioning chloroplasts upon exposure to light. Liu et al. explored the factors that interact to effect this developmental pausing. Previously, the transcription factors EIN3 and PIF3 have each been shown to repress transcription of key photosynthetic genes and chloroplast differentiation. Ethylene accumulates in soil and represses hypocotyl growth, as well as expression of light-responsive genes. The authors show that EIN3 and PIF3 form a “physically-interactive transcription factor pair” that binds to target promoters to repress transcription. Therefore, the combined signals of darkness (which stabilizes PIF3) and ethylene accumulation together suppress the developmental transition from etioplast to chloroplast. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00508
During embryogenesis, plastids arrest their differentiation as etioplasts (characterized by the distinctive prolamellar bodies, PLBs), and remain poised to complete their differentiation into functioning chloroplasts upon exposure to light. Liu et al. explored the factors that interact to effect this developmental pausing. Previously, the transcription factors EIN3 and PIF3 have each been shown to repress transcription of key photosynthetic genes and chloroplast differentiation. Ethylene accumulates in soil and represses hypocotyl growth, as well as expression of light-responsive genes. The authors show that EIN3 and PIF3 form a “physically-interactive transcription factor pair” that binds to target promoters to repress transcription. Therefore, the combined signals of darkness (which stabilizes PIF3) and ethylene accumulation together suppress the developmental transition from etioplast to chloroplast. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00508
Light controls protein localization through phytochrome-mediated alternative promoter selection ($)
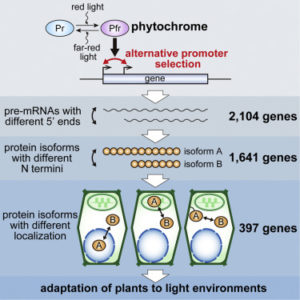 Previous studies have shown that some genes use multiple promoters, but the extent to which this occurs has not been fully resolved. Ushijima et al. showed widespread phytochrome-mediated differential promoter use in response to light. They identified more than 2000 genes with light-dependent alternative promoter selection, most affecting the length of the N-terminus of the protein. From these, they verified nearly 400 in which the different protein isoforms localize differentially within the cell. They further investigate how this affects glycerate kinase (GLYK), which they show is expressed as a longer, plastid-localized protein in the light, but a shorter, cytosolic protein in darkness, providing an cytoplasmic photorespiratory bypass mechanism. (Summary by Mary Williams) Cell 10.1016/j.cell.2017.10.018
Previous studies have shown that some genes use multiple promoters, but the extent to which this occurs has not been fully resolved. Ushijima et al. showed widespread phytochrome-mediated differential promoter use in response to light. They identified more than 2000 genes with light-dependent alternative promoter selection, most affecting the length of the N-terminus of the protein. From these, they verified nearly 400 in which the different protein isoforms localize differentially within the cell. They further investigate how this affects glycerate kinase (GLYK), which they show is expressed as a longer, plastid-localized protein in the light, but a shorter, cytosolic protein in darkness, providing an cytoplasmic photorespiratory bypass mechanism. (Summary by Mary Williams) Cell 10.1016/j.cell.2017.10.018
Chromatin accessibility changes between Arabidopsis stem cells and mesophyll cells illuminate cell type-specific
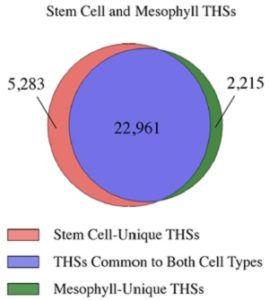 What gives stem cells their plasticity and why are differentiated cells so specialized? Sijacic et al. approach this question by analyzing transcription factor (TF) accessibility to chromatin. Nuclei were isolated from shoot apical meristem (SAM) pluripotent stem cells and fully-differentiated mesophyll cells (using the INTACT technique) to assess chromatin dynamics and transcription factor regulation (ATAC-seq methods). Quantitatively, more regions were accessible in SAM compared to mesophyll nuclei. Of these regions, those in mesophyll nuclei were largely similar to those found in SAM nuclei, but the reverse was not true. The SAM nuclei had more TF binding motifs, likely contributing to stem cell plasticity. The authors further identified TF regulatory networks as well as their downstream components that were unique only to SAM nuclei, highlighting functional differences in each cell type. The data gained here, as well as the techniques used, will be helpful to unraveling the mechanism behind cell fate specification and the role TFs play. (Summary by Alecia Biel) BioRxiv. 10.1101/213900.
What gives stem cells their plasticity and why are differentiated cells so specialized? Sijacic et al. approach this question by analyzing transcription factor (TF) accessibility to chromatin. Nuclei were isolated from shoot apical meristem (SAM) pluripotent stem cells and fully-differentiated mesophyll cells (using the INTACT technique) to assess chromatin dynamics and transcription factor regulation (ATAC-seq methods). Quantitatively, more regions were accessible in SAM compared to mesophyll nuclei. Of these regions, those in mesophyll nuclei were largely similar to those found in SAM nuclei, but the reverse was not true. The SAM nuclei had more TF binding motifs, likely contributing to stem cell plasticity. The authors further identified TF regulatory networks as well as their downstream components that were unique only to SAM nuclei, highlighting functional differences in each cell type. The data gained here, as well as the techniques used, will be helpful to unraveling the mechanism behind cell fate specification and the role TFs play. (Summary by Alecia Biel) BioRxiv. 10.1101/213900.
Cross-species functional diversity within the PIN auxin efflux protein family
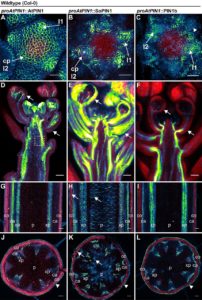 Polar localized PIN FORMED (PIN) efflux carriers proteins organize directional auxin flow and accumulation. Most flowering plants have another family of PIN proteins called Sister of PIN1 (SoPIN1), which Arabidopsis and members of the Brassicacea family do not have. The grass Brachypodium dystachion has one SoPIN1 gene and two PIN1 genes: PIN1a and PIN1b. O´Connor and collaborators created CRISPR mutants in B. dystachion to study the function of these genes. The SoPIN1 mutant showed severe defects, similar to Arabidopsis pin1 mutant. By contrast the Brachypodium pin1a and pin1b mutants showed no clear organ defects. Heterologous expression of SoPIN1 partially complemented the Arabidopsis pin1-613 mutant; conversely, even though PIN1 was highly expressed and polar in the meristems it was not able to mediate organ initiations; showing a clear different function independent of transcriptional control. (Summary by Cecelia Vasquez-Robinet) eLife 10.7554/eLife.31804
Polar localized PIN FORMED (PIN) efflux carriers proteins organize directional auxin flow and accumulation. Most flowering plants have another family of PIN proteins called Sister of PIN1 (SoPIN1), which Arabidopsis and members of the Brassicacea family do not have. The grass Brachypodium dystachion has one SoPIN1 gene and two PIN1 genes: PIN1a and PIN1b. O´Connor and collaborators created CRISPR mutants in B. dystachion to study the function of these genes. The SoPIN1 mutant showed severe defects, similar to Arabidopsis pin1 mutant. By contrast the Brachypodium pin1a and pin1b mutants showed no clear organ defects. Heterologous expression of SoPIN1 partially complemented the Arabidopsis pin1-613 mutant; conversely, even though PIN1 was highly expressed and polar in the meristems it was not able to mediate organ initiations; showing a clear different function independent of transcriptional control. (Summary by Cecelia Vasquez-Robinet) eLife 10.7554/eLife.31804
A plant-specific kinesin KinG regulates intra- and intercellular movement of SHORT-ROOT
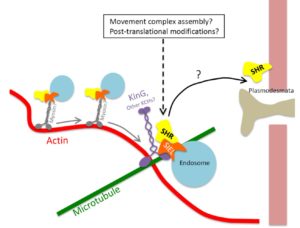 SHORT-ROOT (SRT) is a transcription factor that has previously been shown to move between cells and so contribute to developmental patterning. Spiegelman et al. investigated the cellular machinery that contributes to SRT’s movement. Previous work showed that the movement of SRT depends on the endosome and intact microtubules. In this work the authors identified KinG, a kinesin (microtubule motor protein) that interacts with SRT. Through co-expression in leaf epidermal cells, they observed a siginificant co-localization of SRT and KinG. They also used CRISPR/Cas9 to generate a kinG mutant, and found that SRT movement is reduced in it. They propose a model in which SRT association with KinG promotes pausing, which may facilitate formation of a movement-competent complex. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.17.01518
SHORT-ROOT (SRT) is a transcription factor that has previously been shown to move between cells and so contribute to developmental patterning. Spiegelman et al. investigated the cellular machinery that contributes to SRT’s movement. Previous work showed that the movement of SRT depends on the endosome and intact microtubules. In this work the authors identified KinG, a kinesin (microtubule motor protein) that interacts with SRT. Through co-expression in leaf epidermal cells, they observed a siginificant co-localization of SRT and KinG. They also used CRISPR/Cas9 to generate a kinG mutant, and found that SRT movement is reduced in it. They propose a model in which SRT association with KinG promotes pausing, which may facilitate formation of a movement-competent complex. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.17.01518
SHORTROOT-mediated increase in stomatal density has no impact on photosynthetic efficiency
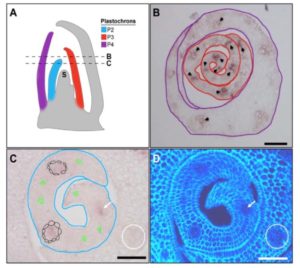 SHORTROOT (SRT) is a transcription factor that contributes to developmental patterning non-cell autonomously, by moving between cells. In leaves, SRT has been shown to contribute to sub-epidermal patterning specified by distance from the vein. Schuler et al. explored whether it also contributes to epidermal patterning, specifically stomatal patterning, in rice leaves. They found that after altering the expression domain of SRT to include bundle-sheath cells surrounding the vein, stomatal patterning was affected. Interestingly, although the plants ectopically expressing SRT produced additional stomata, there was no measurable increase in their photosynthetic efficiency. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.17.01005
SHORTROOT (SRT) is a transcription factor that contributes to developmental patterning non-cell autonomously, by moving between cells. In leaves, SRT has been shown to contribute to sub-epidermal patterning specified by distance from the vein. Schuler et al. explored whether it also contributes to epidermal patterning, specifically stomatal patterning, in rice leaves. They found that after altering the expression domain of SRT to include bundle-sheath cells surrounding the vein, stomatal patterning was affected. Interestingly, although the plants ectopically expressing SRT produced additional stomata, there was no measurable increase in their photosynthetic efficiency. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.17.01005
Arabidopsis perfusion platform RootChip reveals local adaptations of roots
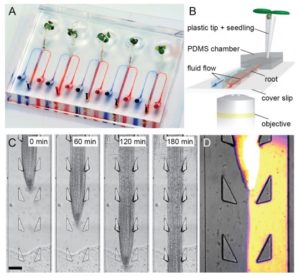 Roots are essential plant organs that need to respond and adapt to dynamic environments. It is still unknown if these adaptations are based on systemic or cell-based responses. Stanley and colleagues created the dual-flow-RootChip, a platform for the heterogeneous perfusion of Arabidopsis roots to investigate the interactions between roots and the environment. They show that there is cell-level adaptation of root hair development under asymmetric phosphate perfusion. This paper demonstrates that Arabidopsis roots can locally adapt to heterogeneous conditions at physiological and transcriptional levels. The dual-flow-RootChip can be used to investigate other root-environment interactions, including calcium signaling or microbe-root relationships. (Summary by Julia Miller) New Phytol. 10.1111/nph.14887
Roots are essential plant organs that need to respond and adapt to dynamic environments. It is still unknown if these adaptations are based on systemic or cell-based responses. Stanley and colleagues created the dual-flow-RootChip, a platform for the heterogeneous perfusion of Arabidopsis roots to investigate the interactions between roots and the environment. They show that there is cell-level adaptation of root hair development under asymmetric phosphate perfusion. This paper demonstrates that Arabidopsis roots can locally adapt to heterogeneous conditions at physiological and transcriptional levels. The dual-flow-RootChip can be used to investigate other root-environment interactions, including calcium signaling or microbe-root relationships. (Summary by Julia Miller) New Phytol. 10.1111/nph.14887
Genetic components of root architecture remodeling in response to salt stress
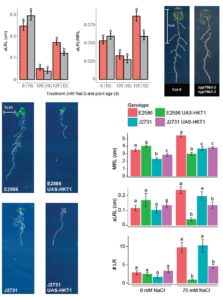
While everyone knows that salt stress reduces root growth, Julkowska and colleagues examined salt-stress induced changes in Root System Architecture (RSA) by studying 347 Arabidopsis accessions. The authors collected 17 RSA traits and developed an app allowing interactive exploration of collected data. GWAS identified 100 loci, among which were CYP79B2 and HKT1. CYP79B2 was validated to be involved in maintenance of lateral root growth during salt stress, possibly by altering auxin biosynthesis. Enhanced expression of the HKT1 ion transporter in the root stele repressed lateral root formation under salt stress conditions, revealing that while retention of salt ions in the root is an excellent mechanism of salinity tolerance in larger plants, it is detrimental for primordia development in young seedlings. The validation of other identified loci remains to be revealed. (Summary by Magdalena Julkowska) Plant Cell 10.1105/tpc.16.00680
Members of the abscisic acid co-receptor PP2C protein family mediate salicylic acid-abscisic acid crosstalk
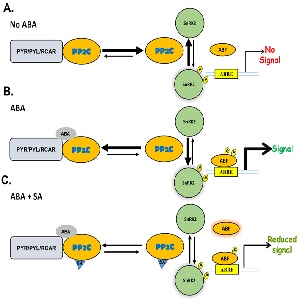 If we know anything about how ABA regulates plant response to environmental stresses it is that the pathway if very complicated and anything but straightforward. The ‘core’ ABA pathway is activated by first inhibiting a specific class of protein phosphatases, PP2Cs. This inhibition of PP2Cs allows for activation of downstream factors and induction of the ABA pathway. PP2Cs have several clades, with each clade having specific functions, regulating both stressed and non-stressed conditions. ABA enhances plant susceptibility to pathogen invasion by suppressing immune responses, , and inhibits salicylic acid(SA)-induced gene expression. SA is a hormone well characterized in enhancing plant immune response whereas ABA suppresses immune response while enhancing plant susceptibility to pathogen invasion. Manohar et al. investigate the trade-off in regulating these two antagonistic hormone pathways: ABA or SA. SA was found to inhibit ABA-induced gene expression. The authors identified a novel SA binding protein in the Clade D PP2Cs that ABA also binds. ABA and SA binding to PP2C results in a game of inhibitions where SA inhibits canonical ABA inhibition of PP2Cs. The data provided supports a model in which SA helps to stabilize PP2Cs, acting against the role of ABA in promoting degradation via proteolysis of PP2Cs. This enhances our understanding of the extensive crosstalk between hormones during environmental stresses while also adding more complexity and variability to what is known to be a very complex process. (Summary by Alecia Biel) Plant Direct. 10.1002/pld3.20.
If we know anything about how ABA regulates plant response to environmental stresses it is that the pathway if very complicated and anything but straightforward. The ‘core’ ABA pathway is activated by first inhibiting a specific class of protein phosphatases, PP2Cs. This inhibition of PP2Cs allows for activation of downstream factors and induction of the ABA pathway. PP2Cs have several clades, with each clade having specific functions, regulating both stressed and non-stressed conditions. ABA enhances plant susceptibility to pathogen invasion by suppressing immune responses, , and inhibits salicylic acid(SA)-induced gene expression. SA is a hormone well characterized in enhancing plant immune response whereas ABA suppresses immune response while enhancing plant susceptibility to pathogen invasion. Manohar et al. investigate the trade-off in regulating these two antagonistic hormone pathways: ABA or SA. SA was found to inhibit ABA-induced gene expression. The authors identified a novel SA binding protein in the Clade D PP2Cs that ABA also binds. ABA and SA binding to PP2C results in a game of inhibitions where SA inhibits canonical ABA inhibition of PP2Cs. The data provided supports a model in which SA helps to stabilize PP2Cs, acting against the role of ABA in promoting degradation via proteolysis of PP2Cs. This enhances our understanding of the extensive crosstalk between hormones during environmental stresses while also adding more complexity and variability to what is known to be a very complex process. (Summary by Alecia Biel) Plant Direct. 10.1002/pld3.20.
Barriers to integration of bioinformatics into undergraduate life sciences education
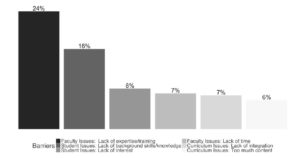 Today’s biology students need to be trained to work with large datasets, meaning that mathematics, statistics and computer science should regularly be integrated into their biology courses. Williams et al. carried out a survey to determine the extent to which this occurs. Although 95% of respondents agree that bioinformatics training is important, 60% do not teach courses with substantial bioinformatics content. The most common reason cited is lack of faculty training. This manuscript lists several training opportunities and also a link to a website where faculty can find these resources. (Summary by Mary Williams) bioRxiv
Today’s biology students need to be trained to work with large datasets, meaning that mathematics, statistics and computer science should regularly be integrated into their biology courses. Williams et al. carried out a survey to determine the extent to which this occurs. Although 95% of respondents agree that bioinformatics training is important, 60% do not teach courses with substantial bioinformatics content. The most common reason cited is lack of faculty training. This manuscript lists several training opportunities and also a link to a website where faculty can find these resources. (Summary by Mary Williams) bioRxiv
Laccaria bicolor MiSSP8 is a small-secreted protein decisive for the establishment of the ectomycorrhizal symbiosis
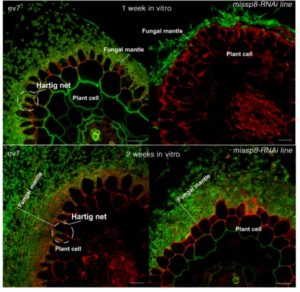 Mycorrhizal symbiosis involves extensive signaling between plants and their fungal partners. Mycorrhiza-induced small secreted proteins (MiSSPs) have been hypothesized to be involved in diverse processes to suppress plant defense and promote fungal life-cycles. Clement et al. functionally characterized a highly expressed small secreted protein MiSSP8 from Laccaria bicolor, and showed that it helps establish ectomycorrhizal symbiosis. MiSSP8-RNAi mutants of L. bicolor are impaired in their mycorrhization activity and do not support the formation of the Hartig net (the network of fungal hyphae that surround the root and extends between cells). When expressed transiently in Nicotiana benthamiana MiSSP8 shows a co-localization with plasmodesmata. This work suggests that MiSSP8 has a possible dual role, the first in regulating hyphal formation and a second role related to plasmodesmata localization which remains to be elucidated. (Summary by Amey Redkar) bioRxiv. 10.1101/218131
Mycorrhizal symbiosis involves extensive signaling between plants and their fungal partners. Mycorrhiza-induced small secreted proteins (MiSSPs) have been hypothesized to be involved in diverse processes to suppress plant defense and promote fungal life-cycles. Clement et al. functionally characterized a highly expressed small secreted protein MiSSP8 from Laccaria bicolor, and showed that it helps establish ectomycorrhizal symbiosis. MiSSP8-RNAi mutants of L. bicolor are impaired in their mycorrhization activity and do not support the formation of the Hartig net (the network of fungal hyphae that surround the root and extends between cells). When expressed transiently in Nicotiana benthamiana MiSSP8 shows a co-localization with plasmodesmata. This work suggests that MiSSP8 has a possible dual role, the first in regulating hyphal formation and a second role related to plasmodesmata localization which remains to be elucidated. (Summary by Amey Redkar) bioRxiv. 10.1101/218131
Mycorrhizal fungi shaped the evolution of terrestrial plants
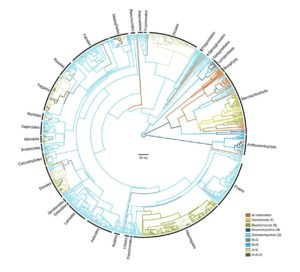 Mutualistic associations between plants and fungi are incredibly widespread, occurring in 90% of extant land plants, and likely are the most ecologically important symbiotic relationships on Earth. Fungi played an integral part in land plant evolution; roots only evolved after early land plants colonised terrestrial environments and these associations allowed for mutualistic exchange of inorganic nutrients and water for carbohydrates in harsh environments predating existing soils. The majority of extant land plants are associated with the Mucoromycota subphylum Glomeromycotina, a division of arbuscular mycorrhizal fungi that penetrate the cortical cells of plant roots for nutrient exchange. Because of this associative abundance together with fossil evidence, it has long been hypothesised that this division of fungi facilitated colonisation of land by early plants. However, a recent study showed evidence that another Mucoromycota subphylum, Mucoromycotina, was likely responsible for enabling the establishment of early land plants, and that Glomeromycotina was a secondary acquisition. This study also revealed that transitions from one fungal phyla to another, such as to species from the Basidiomycota and Ascomycota, or forming symbioses with multiple fungal partners, may have led to unique niche expansions and radiations such as those seen in the orchids, the largest family of land plants on Earth, as well as plants adapted to nutrient poor or acidic soil. The importance of Mucoromycotina in land plant evolution will likely become more apparent as the relationships between mycorrhizal fungi and plants are further explored. (Summary by Danielle Roodt Prinsloo) bioRxiv 10.1101/213090 (Cover illustration by Perry Shirley)
Mutualistic associations between plants and fungi are incredibly widespread, occurring in 90% of extant land plants, and likely are the most ecologically important symbiotic relationships on Earth. Fungi played an integral part in land plant evolution; roots only evolved after early land plants colonised terrestrial environments and these associations allowed for mutualistic exchange of inorganic nutrients and water for carbohydrates in harsh environments predating existing soils. The majority of extant land plants are associated with the Mucoromycota subphylum Glomeromycotina, a division of arbuscular mycorrhizal fungi that penetrate the cortical cells of plant roots for nutrient exchange. Because of this associative abundance together with fossil evidence, it has long been hypothesised that this division of fungi facilitated colonisation of land by early plants. However, a recent study showed evidence that another Mucoromycota subphylum, Mucoromycotina, was likely responsible for enabling the establishment of early land plants, and that Glomeromycotina was a secondary acquisition. This study also revealed that transitions from one fungal phyla to another, such as to species from the Basidiomycota and Ascomycota, or forming symbioses with multiple fungal partners, may have led to unique niche expansions and radiations such as those seen in the orchids, the largest family of land plants on Earth, as well as plants adapted to nutrient poor or acidic soil. The importance of Mucoromycotina in land plant evolution will likely become more apparent as the relationships between mycorrhizal fungi and plants are further explored. (Summary by Danielle Roodt Prinsloo) bioRxiv 10.1101/213090 (Cover illustration by Perry Shirley)
Ecosystem responses to elevated CO2 are governed by plant-soil interactions and the cost of nitrogen acquisition
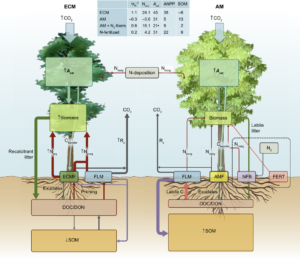 How does the cost of nitrogen acquisition affect how an ecosystem responds to elevated CO2? Terrer et al. have addressed this question in a comprehensive review of findings from elevated CO2 experiments, using a plant economics framework. The authors describe ecosystem responses, particularly those of plants to elevated CO2, by interactions with mycorrhizal fungi and symbiotic N-fixing microbes, and explain these responses as a relationship between costs and benefits. These established interactions that allow N-acquisition positively correlate with plant growth and photosynthetic capacity and have a negative co-relation with carbon storage. The review summarizes that the predicted models which aim to improve the carbon feedback loop as a result of elevated CO2 emissions may be improved by treating N as a resource for plants that can be acquired at the expense of enregy, and will have lower costs depending upon the status of the plant-microbe symbiosis. (Summary by Amey Redkar) New Phytol. 10.1111/nph.14872/full
How does the cost of nitrogen acquisition affect how an ecosystem responds to elevated CO2? Terrer et al. have addressed this question in a comprehensive review of findings from elevated CO2 experiments, using a plant economics framework. The authors describe ecosystem responses, particularly those of plants to elevated CO2, by interactions with mycorrhizal fungi and symbiotic N-fixing microbes, and explain these responses as a relationship between costs and benefits. These established interactions that allow N-acquisition positively correlate with plant growth and photosynthetic capacity and have a negative co-relation with carbon storage. The review summarizes that the predicted models which aim to improve the carbon feedback loop as a result of elevated CO2 emissions may be improved by treating N as a resource for plants that can be acquired at the expense of enregy, and will have lower costs depending upon the status of the plant-microbe symbiosis. (Summary by Amey Redkar) New Phytol. 10.1111/nph.14872/full