What We’re Reading: May 25th
Review: A guide to sequence your favorite plant genomes
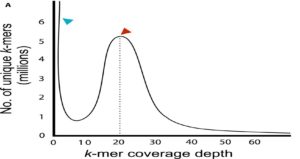 Every year, genome assembly gets faster and cheaper. Li and Harkness provide a practical guide to today’s methods, with caveats and precautions that need to be considered at each step, even questions to ask before embarking on a sequencing project. Topics include how to estimate genome size and complexity, pros and cons of different sequencing platforms, and best practices for genome assembly. The authors also recommend that you check with the Open Green Genomes Initiative (OGG) and 10,000 Plants Genome Sequencing Project (10KP) to see if your genome of interest is already being sequenced. (Summary by Mary Williams) App. Plant Sci. 10.1002/aps3.1030
Every year, genome assembly gets faster and cheaper. Li and Harkness provide a practical guide to today’s methods, with caveats and precautions that need to be considered at each step, even questions to ask before embarking on a sequencing project. Topics include how to estimate genome size and complexity, pros and cons of different sequencing platforms, and best practices for genome assembly. The authors also recommend that you check with the Open Green Genomes Initiative (OGG) and 10,000 Plants Genome Sequencing Project (10KP) to see if your genome of interest is already being sequenced. (Summary by Mary Williams) App. Plant Sci. 10.1002/aps3.1030
Opinion: Non-mycorrhizal Plants: The exceptions that prove the rule
 Most vascular plants establish in their roots a multifunctional symbiosis with arbuscular mycorrhizal (AM) fungi. Among the 29% that do not host AM fungi are 66-92% of the members of the Brassicaceae family, including some major crops (broccoli, oilseed rape) as well as the model plant Arabidopsis thaliana. Although Arabidopsis is considered a non-host, conditional and rudimentary AM colonization has been described in it. In this review, Cosme et al. indicate that the existing (and extensive) genomic information from the Brassicaceae family may shed light on the factors that drive AM symbiotic incompatibility, based on the presence or absence of symbiotic toolkit genes (NFP, DMI2, DMI3, CASTOR and others) required for colonization and establishment of the arbuscules. (Summary by Isabel Mendoza) Trends Plant Sci. 10.1016/j.tplants.2018.04.004
Most vascular plants establish in their roots a multifunctional symbiosis with arbuscular mycorrhizal (AM) fungi. Among the 29% that do not host AM fungi are 66-92% of the members of the Brassicaceae family, including some major crops (broccoli, oilseed rape) as well as the model plant Arabidopsis thaliana. Although Arabidopsis is considered a non-host, conditional and rudimentary AM colonization has been described in it. In this review, Cosme et al. indicate that the existing (and extensive) genomic information from the Brassicaceae family may shed light on the factors that drive AM symbiotic incompatibility, based on the presence or absence of symbiotic toolkit genes (NFP, DMI2, DMI3, CASTOR and others) required for colonization and establishment of the arbuscules. (Summary by Isabel Mendoza) Trends Plant Sci. 10.1016/j.tplants.2018.04.004
CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation
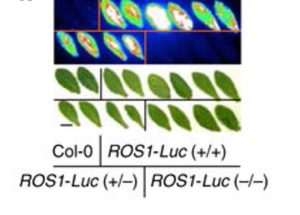 Genome modification is a powerful tool, yet precise genome engineering in plants remains challenging. Miki et al. report on the sequential transformation method for gene targeting in Arabidopsis. The parental plants expressing CRISPR/Cas9 under the egg cell- and early embryo-specific DD45 promoter were transformed with constructs containing (1) a homology-directed repair donor sequence, (2) sgRNA targeting the locus of interest and (3) a selectable marker. Using this technique, the group made functional GFP and luciferase fusions with native genes that were successfully tagged at the 5’ and 3’ end. The described technique is also suitable to induce non-synonymous mutations in the coding sequence, without any additional mutations. In future the authors hope to improve the efficiency of this gene targeting technique above the reported 5-10%. (Summary by Magdalena Julkowska) Nature Comms. 10.1038/s41467-018-04416-0
Genome modification is a powerful tool, yet precise genome engineering in plants remains challenging. Miki et al. report on the sequential transformation method for gene targeting in Arabidopsis. The parental plants expressing CRISPR/Cas9 under the egg cell- and early embryo-specific DD45 promoter were transformed with constructs containing (1) a homology-directed repair donor sequence, (2) sgRNA targeting the locus of interest and (3) a selectable marker. Using this technique, the group made functional GFP and luciferase fusions with native genes that were successfully tagged at the 5’ and 3’ end. The described technique is also suitable to induce non-synonymous mutations in the coding sequence, without any additional mutations. In future the authors hope to improve the efficiency of this gene targeting technique above the reported 5-10%. (Summary by Magdalena Julkowska) Nature Comms. 10.1038/s41467-018-04416-0
Gravisensors in plant cells behave like an active granular liquid
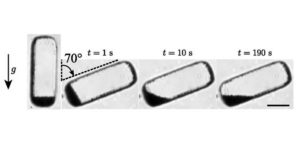 When you try to tip rocks out of a bucket, nothing happens until the steepness of the angle is sufficient to start them shifting (the “avalanche angle”). By contrast, water starts moving with even the slightest tilt to the bucket. Now think about the gravity-sensing particles in plant cells, starch-rich statoliths, which are quite large relative to the cells that contain them. Bérut et al. asked how plant gravity responses can be so fine when their gravity sensing machinery appears so course. By observing the behaviour of lots and lots of statoliths, the authors concluded that statoliths act more like water than rocks; or actually, like rocks that are being agitated, causing them to unjam and behave like a fluid. Using a biomimetic approach, the authors ruled out thermal or Brownian sources, and conclude that this agitation stems from cell activity, specifically the dynamics of the actin cytoskeleton, consistent with previous studies using inhibitors of the actin cytoskeleton. In addition to really cool findings, this paper has terrific videos! (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1801895115
When you try to tip rocks out of a bucket, nothing happens until the steepness of the angle is sufficient to start them shifting (the “avalanche angle”). By contrast, water starts moving with even the slightest tilt to the bucket. Now think about the gravity-sensing particles in plant cells, starch-rich statoliths, which are quite large relative to the cells that contain them. Bérut et al. asked how plant gravity responses can be so fine when their gravity sensing machinery appears so course. By observing the behaviour of lots and lots of statoliths, the authors concluded that statoliths act more like water than rocks; or actually, like rocks that are being agitated, causing them to unjam and behave like a fluid. Using a biomimetic approach, the authors ruled out thermal or Brownian sources, and conclude that this agitation stems from cell activity, specifically the dynamics of the actin cytoskeleton, consistent with previous studies using inhibitors of the actin cytoskeleton. In addition to really cool findings, this paper has terrific videos! (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1801895115
Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity ($)
 Like all plant hormones, ABA controls diverse processes, including stomatal aperture and seed dormancy, as well as growth rate. Miao et al. have used CRISPR/Cas9 technology in rice to selectively mutate subsets of the 13-member PYL family of ABA receptors. They found notable differences when mutating group I (PYL1–PYL6 and PYL12) versus group II (PYL7–PYL11 and PYL13) PYL genes, including differences in growth rate, heading date, and seed dormancy. Notably, in field conditions, the triple mutant pyl1/4/6 had 25% or 31% increased grain yield as compared to wild type – an exciting result. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1804774115
Like all plant hormones, ABA controls diverse processes, including stomatal aperture and seed dormancy, as well as growth rate. Miao et al. have used CRISPR/Cas9 technology in rice to selectively mutate subsets of the 13-member PYL family of ABA receptors. They found notable differences when mutating group I (PYL1–PYL6 and PYL12) versus group II (PYL7–PYL11 and PYL13) PYL genes, including differences in growth rate, heading date, and seed dormancy. Notably, in field conditions, the triple mutant pyl1/4/6 had 25% or 31% increased grain yield as compared to wild type – an exciting result. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1804774115
HRB2-BBX21 interaction regulates stomatal aperture
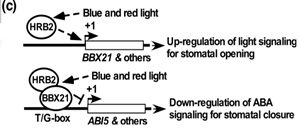 Stomata must maintain a balance between opening for carbon uptake and closing to prevent dehydration and pathogen invasion during times of stress. Kang et al. investigate this relationship, utilizing light and ABA signaling to modulate stomatal dynamics. HBR2 was identified as a chromatin-remodeling factor, corresponding to the gene PKL. The mutant hrb2 has defects in hypocotyl growth under light conditions similar to that of pkl-1 (tested in this work) and other pkl alleles (referenced). In the absence of ABA, hbr2 had reduced stomatal apertures, whereas bbx21-1 mutants were WT-like. BBX21 is a B-box protein involved in various light responses. Both mutants showed reduced stomatal apertures in the presence of ABA. Furthermore, HRB2 upregulates the expression of BBX21, which is light inducible. To further elucidate the role of HRB2 in the ABA pathway, expression of the ABA signaling gene ABI5 was investigated and found to be upregulated in hbr2, pkl-1, and bbx21-1 leaves. Overexpression of ABI5 reduces stomatal apertures and may account for the phenotype of hrb2. BBX21 was found to associate with the ABI5 promoter. This leads to a model in which BBX21 binds to the ABI5 promoter and recruits HRB2 to modulate chromatin at this locus (Summary by Alecia Biel) Plant Cell Environ. 10.1111/pce.13336.
Stomata must maintain a balance between opening for carbon uptake and closing to prevent dehydration and pathogen invasion during times of stress. Kang et al. investigate this relationship, utilizing light and ABA signaling to modulate stomatal dynamics. HBR2 was identified as a chromatin-remodeling factor, corresponding to the gene PKL. The mutant hrb2 has defects in hypocotyl growth under light conditions similar to that of pkl-1 (tested in this work) and other pkl alleles (referenced). In the absence of ABA, hbr2 had reduced stomatal apertures, whereas bbx21-1 mutants were WT-like. BBX21 is a B-box protein involved in various light responses. Both mutants showed reduced stomatal apertures in the presence of ABA. Furthermore, HRB2 upregulates the expression of BBX21, which is light inducible. To further elucidate the role of HRB2 in the ABA pathway, expression of the ABA signaling gene ABI5 was investigated and found to be upregulated in hbr2, pkl-1, and bbx21-1 leaves. Overexpression of ABI5 reduces stomatal apertures and may account for the phenotype of hrb2. BBX21 was found to associate with the ABI5 promoter. This leads to a model in which BBX21 binds to the ABI5 promoter and recruits HRB2 to modulate chromatin at this locus (Summary by Alecia Biel) Plant Cell Environ. 10.1111/pce.13336.
The cytokinin trans-Zeatin plays a role in long distance nitrogen signaling ($)
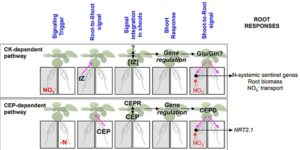 Plants modify gene expression and physiological processes to overcome temporal and spatial variations in nitrogen availability. These modifications rely on complex root-shoot-root signaling networks which are triggered by cytokinin biosynthesis. Poitout and colleagues use mutant analysis, transcriptome analysis and hormone profiling to understand complex long distance nitrogen signaling networks in Arabidopsis thaliana. In the shoots, the active form of the cytokinin trans-Zeatin plays a role in the integration of rapid gene regulation and long-term acclimation to heterogenous NO3– supply. Additionally, glutamate/glutamine metabolism seems to be a target of trans-Zeatin root-to-shoot translocation. There are also trans-Zeatin-independent pathways that regulate shoot gene expression and NO3– uptake. This paper elucidates the role of cytokinins in nitrogen signaling and aids to integrate nitrogen-related long distance signaling networks. (Summary by Julia Miller) Plant Cell 10.1105/tpc.18.00011
Plants modify gene expression and physiological processes to overcome temporal and spatial variations in nitrogen availability. These modifications rely on complex root-shoot-root signaling networks which are triggered by cytokinin biosynthesis. Poitout and colleagues use mutant analysis, transcriptome analysis and hormone profiling to understand complex long distance nitrogen signaling networks in Arabidopsis thaliana. In the shoots, the active form of the cytokinin trans-Zeatin plays a role in the integration of rapid gene regulation and long-term acclimation to heterogenous NO3– supply. Additionally, glutamate/glutamine metabolism seems to be a target of trans-Zeatin root-to-shoot translocation. There are also trans-Zeatin-independent pathways that regulate shoot gene expression and NO3– uptake. This paper elucidates the role of cytokinins in nitrogen signaling and aids to integrate nitrogen-related long distance signaling networks. (Summary by Julia Miller) Plant Cell 10.1105/tpc.18.00011
Root-expressed phytochrome B1 and B2 regulate shoot growth in nature
 Light is not only a driving force of photosynthesis, but also an important environmental signal. Oh et al. report that a number of light sensing phytochromes are expressed in roots of native tobacco (N. attenuata) at higher levels than in the shoots. The authors investigated the roles of those root phytochromes by grafting light-blind roots with light-sensitive scions. The plants with roots lacking phytochrome B1 and B2 showed delayed elongation of the hypocotyl under controlled and field conditions. Interestingly, this effect was dependent on temperature. The authors suggest that the root-specific phytochromes serve to sense aboveground light and temperature, and affect aboveground plant growth through root-to-shoot signaling. How the light-sensing roots affect plant’s fitness remains to be determined. (Summary by Magdalena Julkowska) Plant Cell Environ. 10.1111/pce.13341
Light is not only a driving force of photosynthesis, but also an important environmental signal. Oh et al. report that a number of light sensing phytochromes are expressed in roots of native tobacco (N. attenuata) at higher levels than in the shoots. The authors investigated the roles of those root phytochromes by grafting light-blind roots with light-sensitive scions. The plants with roots lacking phytochrome B1 and B2 showed delayed elongation of the hypocotyl under controlled and field conditions. Interestingly, this effect was dependent on temperature. The authors suggest that the root-specific phytochromes serve to sense aboveground light and temperature, and affect aboveground plant growth through root-to-shoot signaling. How the light-sensing roots affect plant’s fitness remains to be determined. (Summary by Magdalena Julkowska) Plant Cell Environ. 10.1111/pce.13341
Identification of cold tolerance genes and a functional allele that confers cold tolerance ($)
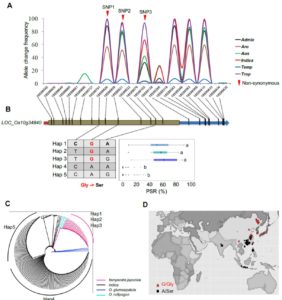 Xiao et al. used 1,033 rice accessions for GWAS to identify QTLs associated with cold tolerance. In general, japonica-type varieties show greater cold tolerance than indica types. The authors identified many QTLs for cold tolerance at the seedling and booting (initiation of panicles) stages. They identified a locus correlated with cold tolerance, and a single-nucleotide polymorphism that is differentially present in high- versus low-altitude accessions. Further analysis including transgenic introduction or silencing of the candidate gene carrying the cold-tolerance associated SNP demonstrated its role in cold tolerance. The gene encodes a protein with a pectin lyase fold, annotated to be involved in cell wall metabolism. The authors speculate that the pectin content of cell walls, as regulated by this gene, could affect cold tolerance in rice. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00209
Xiao et al. used 1,033 rice accessions for GWAS to identify QTLs associated with cold tolerance. In general, japonica-type varieties show greater cold tolerance than indica types. The authors identified many QTLs for cold tolerance at the seedling and booting (initiation of panicles) stages. They identified a locus correlated with cold tolerance, and a single-nucleotide polymorphism that is differentially present in high- versus low-altitude accessions. Further analysis including transgenic introduction or silencing of the candidate gene carrying the cold-tolerance associated SNP demonstrated its role in cold tolerance. The gene encodes a protein with a pectin lyase fold, annotated to be involved in cell wall metabolism. The authors speculate that the pectin content of cell walls, as regulated by this gene, could affect cold tolerance in rice. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00209
The repetitive effector Rsp3 promotes the virulence of the corn smut fungus Ustilago maydis
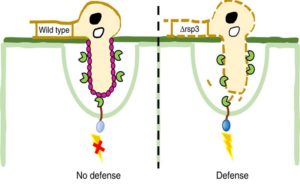 The corn smut fungus Ustilago maydis manipulates maize tissues and cells through the secretion of effectors that modulate host protein activities. In a recent article published in Nature Communications, Ma et al. characterize a highly repetitive effector protein family (Rsp3, repetitive secreted protein 3) that significantly contributes to fungal virulence. The transcriptional induction of Rsp3 coincided with the establishment of intracellular colonization while Rsp3 protein was shown to decorate fungal hyphae and interact with host-secreted anti-fungal proteins (AFPs). Importantly, U. maydis knockout strains lacking the Rsp3 effector displayed reduced virulence on wild-type maize seedlings, but remained fully virulent on AFP-silenced host plants. Together these results highlight the role of Rsp3 in binding and preventing the anti-fungal activity of host AFPs during intracellular colonization. Furthermore, the authors demonstrated the widespread prevalence of Rsp3 homologs in several smut fungi and validated the functional conservation of an Rsp3 homolog from an additional maize-infecting pathogen. This work suggests that Rsp3 proteins promote pathogen growth by protecting fungal hyphae from the activity of anti-fungal host proteins. (Summary by Phil Carella) Nature Comms. 10.1038/s41467-018-04149-0
The corn smut fungus Ustilago maydis manipulates maize tissues and cells through the secretion of effectors that modulate host protein activities. In a recent article published in Nature Communications, Ma et al. characterize a highly repetitive effector protein family (Rsp3, repetitive secreted protein 3) that significantly contributes to fungal virulence. The transcriptional induction of Rsp3 coincided with the establishment of intracellular colonization while Rsp3 protein was shown to decorate fungal hyphae and interact with host-secreted anti-fungal proteins (AFPs). Importantly, U. maydis knockout strains lacking the Rsp3 effector displayed reduced virulence on wild-type maize seedlings, but remained fully virulent on AFP-silenced host plants. Together these results highlight the role of Rsp3 in binding and preventing the anti-fungal activity of host AFPs during intracellular colonization. Furthermore, the authors demonstrated the widespread prevalence of Rsp3 homologs in several smut fungi and validated the functional conservation of an Rsp3 homolog from an additional maize-infecting pathogen. This work suggests that Rsp3 proteins promote pathogen growth by protecting fungal hyphae from the activity of anti-fungal host proteins. (Summary by Phil Carella) Nature Comms. 10.1038/s41467-018-04149-0
Plant exosomes mediate cross-kingdom RNA interference ($)
 Small RNAs (sRNAs) trafficking between host plant and pathogens can occur in a wide range of interactions. Pathogen-produced sRNAs have been shown to suppress host plant immunity, and conversely host plant-derived sRNAs can reduce pathogen virulence by targeting pathogenic genes of the pest. Nevertheless, mechanisms ensuring host/pest sRNAs transfer remain unclear. In this study, Cai et al. used the Arabidopsis/Botrytis cinerea pathosystem to identify the molecular vehicle responsible for directional plant sRNA exchange. They first confirm that plant endogenous sRNAs are not only transferred into the fungus, but supress fungal pathogenicity by silencing virulence genes. They further show that plant sRNAs accumulate into exosome-like extracellular vesicles secreted by Arabidopsis cells at infection sites, and later detected in the fungal cells where they are believed to deliver the sRNAs targeting virulence genes. Together, this study describes plant exosomes as the molecular vehicle mediating plant-pathogen sRNAs trafficking, and offers new perspectives in plant disease control. (Summary by Matthias Benoit) Science 10.1126/science.aar4142
Small RNAs (sRNAs) trafficking between host plant and pathogens can occur in a wide range of interactions. Pathogen-produced sRNAs have been shown to suppress host plant immunity, and conversely host plant-derived sRNAs can reduce pathogen virulence by targeting pathogenic genes of the pest. Nevertheless, mechanisms ensuring host/pest sRNAs transfer remain unclear. In this study, Cai et al. used the Arabidopsis/Botrytis cinerea pathosystem to identify the molecular vehicle responsible for directional plant sRNA exchange. They first confirm that plant endogenous sRNAs are not only transferred into the fungus, but supress fungal pathogenicity by silencing virulence genes. They further show that plant sRNAs accumulate into exosome-like extracellular vesicles secreted by Arabidopsis cells at infection sites, and later detected in the fungal cells where they are believed to deliver the sRNAs targeting virulence genes. Together, this study describes plant exosomes as the molecular vehicle mediating plant-pathogen sRNAs trafficking, and offers new perspectives in plant disease control. (Summary by Matthias Benoit) Science 10.1126/science.aar4142



