What We’re Reading: March 3
Update: Stomatal biology of CAM plants
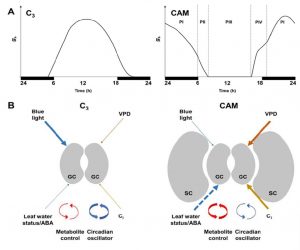 Crassulacean acid metabolism (CAM) plants open their stomata at night, decreasing water loss and increasing water-use efficiency as well as drought tolerance. Males and Griffiths review the stomatal biology of CAM plants as compared to C3 plants. For example, CAM stomata are relatively insensitive to blue light, which is a powerful opening stimulus for C3 stomata; conversely, CAM stomata appear to be more sensitive to plant water status and intercellular CO2 levels than C3 plants. However, deliniating differences between C3 and CAM stomatal biology is complicated by the fact that CAM is a convergent trait that has evolved independently many times in diverse lineages. Facultative CAM plants, which can switch from C3 to CAM when conditions warrant, are promising models. A key objective of these studies is to investigate the possibility of engineering facultative CAM into C3 plants to enable them to weather periods of drought. Plant Physiol. 10.1104/pp.16.01644 Tags: Biochemistry, Bioenergetics, Botany, Ecophysiology, Evolution, Metabolism, Physiology
Crassulacean acid metabolism (CAM) plants open their stomata at night, decreasing water loss and increasing water-use efficiency as well as drought tolerance. Males and Griffiths review the stomatal biology of CAM plants as compared to C3 plants. For example, CAM stomata are relatively insensitive to blue light, which is a powerful opening stimulus for C3 stomata; conversely, CAM stomata appear to be more sensitive to plant water status and intercellular CO2 levels than C3 plants. However, deliniating differences between C3 and CAM stomatal biology is complicated by the fact that CAM is a convergent trait that has evolved independently many times in diverse lineages. Facultative CAM plants, which can switch from C3 to CAM when conditions warrant, are promising models. A key objective of these studies is to investigate the possibility of engineering facultative CAM into C3 plants to enable them to weather periods of drought. Plant Physiol. 10.1104/pp.16.01644 Tags: Biochemistry, Bioenergetics, Botany, Ecophysiology, Evolution, Metabolism, Physiology
Review: Source-sink interactions in plants ($)
 With populations increasing globally, improving crop yield potential is one of the major challenges to the plant biologist, complicated by the changing climate. A better understanding of the source (material producer or exporter, e.g., leaves) – sink (material importer or consumer, e.g., roots, growing fruits and seeds) relationship is very important to develop crop plants with higher yield potential. In this review, Chang and Zhu summarize the concepts and terminology of source-sink interactions and discuss current progress in modern molecular systems biology to manipulate the source-sink interactions in crop plants. The authors also propose a new, integrative systems approach which may help to link molecular mechanisms directly to physiological source-sink interactions. They concluded that “a well modularized and highly mechanistic model holds great potential to help predict yield for different crops under different conditions“. (Summary by Mather Khan) J. Exp. Bot. 10.1093/jxb/erx002 Tags: Physiology, Systems Biology, Transport
With populations increasing globally, improving crop yield potential is one of the major challenges to the plant biologist, complicated by the changing climate. A better understanding of the source (material producer or exporter, e.g., leaves) – sink (material importer or consumer, e.g., roots, growing fruits and seeds) relationship is very important to develop crop plants with higher yield potential. In this review, Chang and Zhu summarize the concepts and terminology of source-sink interactions and discuss current progress in modern molecular systems biology to manipulate the source-sink interactions in crop plants. The authors also propose a new, integrative systems approach which may help to link molecular mechanisms directly to physiological source-sink interactions. They concluded that “a well modularized and highly mechanistic model holds great potential to help predict yield for different crops under different conditions“. (Summary by Mather Khan) J. Exp. Bot. 10.1093/jxb/erx002 Tags: Physiology, Systems Biology, Transport
Review: The cryptic chemical traits that mediate plant community composition
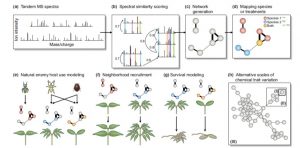 Plants produce a huge variety of specialized metabolites, many with roles in defense. Metabolic profiles rarely follow phylogenetic lines; in fact, closely related species often produce dramatically different suites of metabolites. When it comes to defense chemistry, it is advantageous to be different from your neighbor; species can carve out their own niche as a consequence of the pests and pathogens they can avoid. Sedio explores the use of comparative structural metabolomics to understand the role of specialized metabolites in community ecology. Specifically, he describes an approach that compares the structural similarity of unknown compounds from complex samples. By comparing thousands of structures from hundreds of plants, it should be possible to test the hypothesis that chemical diversity drives ecological coexistence of plant species diversity. New Phytol. 10.1111/nph.14438 Tags: Biotic Interactions, Ecophysiology, Evolution, Metabolism,
Plants produce a huge variety of specialized metabolites, many with roles in defense. Metabolic profiles rarely follow phylogenetic lines; in fact, closely related species often produce dramatically different suites of metabolites. When it comes to defense chemistry, it is advantageous to be different from your neighbor; species can carve out their own niche as a consequence of the pests and pathogens they can avoid. Sedio explores the use of comparative structural metabolomics to understand the role of specialized metabolites in community ecology. Specifically, he describes an approach that compares the structural similarity of unknown compounds from complex samples. By comparing thousands of structures from hundreds of plants, it should be possible to test the hypothesis that chemical diversity drives ecological coexistence of plant species diversity. New Phytol. 10.1111/nph.14438 Tags: Biotic Interactions, Ecophysiology, Evolution, Metabolism,
Technical Advance: Distribution of thylakoid membrane lipids among individual cells of maize leaf ($)
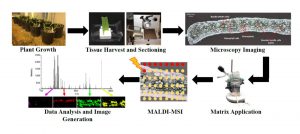 The distribution of metabolites in tissues can be determined in situ through the technique Mass spectrometry imaging (MSI) with matrix-assisted laser desorption ionization (MALDI) (also known as MALDI-MSI). Duenas et al. used MALDI-MSI to analyze the distribution of thylakoid membrane lipids in maize, specifically comparing those of the the bundle sheath cells and mesophyll cells. Maize has Kranz anatomy in which the chloroplasts of the mesophyll cells are specialized for light harvesting, and those of the bundle sheath cells are specialized for the Calvin-Benson Cycle. The authors found that SQDG (sulfoquinovosyldiacylglycerol) species are uniformly distributed in both photosynthetic cell types. By contrast, phosphatidyglycerol (PG) species show cell-specific localization. As an example, 16:0-containing PGs are mostly in bundle-sheath cells whereas 16:1-containing PGs are primarily in mesophyll cells and accumulate as the tissue develops and becomes photosynthetically active. These data demonstrate the power of MALDI-MSI for probing metabolism at the single-cell level. Plant J. 10.1111/tpj.13422 Tags: Biochemistry, Bioenergetics, Metabolism, Physiology
The distribution of metabolites in tissues can be determined in situ through the technique Mass spectrometry imaging (MSI) with matrix-assisted laser desorption ionization (MALDI) (also known as MALDI-MSI). Duenas et al. used MALDI-MSI to analyze the distribution of thylakoid membrane lipids in maize, specifically comparing those of the the bundle sheath cells and mesophyll cells. Maize has Kranz anatomy in which the chloroplasts of the mesophyll cells are specialized for light harvesting, and those of the bundle sheath cells are specialized for the Calvin-Benson Cycle. The authors found that SQDG (sulfoquinovosyldiacylglycerol) species are uniformly distributed in both photosynthetic cell types. By contrast, phosphatidyglycerol (PG) species show cell-specific localization. As an example, 16:0-containing PGs are mostly in bundle-sheath cells whereas 16:1-containing PGs are primarily in mesophyll cells and accumulate as the tissue develops and becomes photosynthetically active. These data demonstrate the power of MALDI-MSI for probing metabolism at the single-cell level. Plant J. 10.1111/tpj.13422 Tags: Biochemistry, Bioenergetics, Metabolism, Physiology
Field-based high throughput phenotyping identifies genes controlling yield in rice
 The classic art of plant breeding involves carefully examining a genetically segregating population for traits of interest. Increasingly, high-throughput, automated phenotyping systems are being used; for example, robots can carry plants to imaging chambers for data collection. However, growth-chamber grown plants do not fully replicate the range of conditions experienced by field-grown plants. Tanger et al. describe the application of a field-based, high throughput phenotyping system for rice (based on one developed for cotton). The tractor-based system uses multispectral reflectance and ultrasonic sensors to measure canopy height, temperature depression, and several reflective ratios at a rate of 3000 plots/hr, as compared to 45 plots/hr for manual phenotyping. As a proof-of-concept, the authors identified four alleles with a negative effect on grain yield that are fixed in a modern cultivar (IR64); replacing these negative alleles should lead to yield improvements. Sci. Rep. 10.1038/srep42839 Tags: Applied Plant Biology, Computational Biology, Genetics, Genomics, Physiology
The classic art of plant breeding involves carefully examining a genetically segregating population for traits of interest. Increasingly, high-throughput, automated phenotyping systems are being used; for example, robots can carry plants to imaging chambers for data collection. However, growth-chamber grown plants do not fully replicate the range of conditions experienced by field-grown plants. Tanger et al. describe the application of a field-based, high throughput phenotyping system for rice (based on one developed for cotton). The tractor-based system uses multispectral reflectance and ultrasonic sensors to measure canopy height, temperature depression, and several reflective ratios at a rate of 3000 plots/hr, as compared to 45 plots/hr for manual phenotyping. As a proof-of-concept, the authors identified four alleles with a negative effect on grain yield that are fixed in a modern cultivar (IR64); replacing these negative alleles should lead to yield improvements. Sci. Rep. 10.1038/srep42839 Tags: Applied Plant Biology, Computational Biology, Genetics, Genomics, Physiology
Genome-wide prediction of metabolic enzymes, pathways and gene clusters in plants
 There remains a considerable knowledge gap between the plant genome and the plant metabolome. To address this, Schläpfer et al. have developed a computational pipeline to identify metabolic enzymes, pathways, and gene clusters. Although metabolic genes are known to cluster in bacteria and fungi, until recently this phenomenon was not known to be widespread in plants. However, in just 18 species, the authors identified nearly 12,000 metabolic gene clusters. Many of these clusters include enzymes that produce a metabolite scaffold and enzymes that modify it. Their data also suggest that local gene duplication has contributed to the diversification of metabolic gene clusters. All the data can be browsed online at www.plantcyc.org. As the authors state, these tools can be used for “systematically cataloging the chemical diversity of plants and revealing Nature’s pharmacopeia in the green world.” Plant Physiol. 10.1104/pp.16.01942 Tags: Biochemistry, Botany, Computational Biology, Genomics, Metabolism
There remains a considerable knowledge gap between the plant genome and the plant metabolome. To address this, Schläpfer et al. have developed a computational pipeline to identify metabolic enzymes, pathways, and gene clusters. Although metabolic genes are known to cluster in bacteria and fungi, until recently this phenomenon was not known to be widespread in plants. However, in just 18 species, the authors identified nearly 12,000 metabolic gene clusters. Many of these clusters include enzymes that produce a metabolite scaffold and enzymes that modify it. Their data also suggest that local gene duplication has contributed to the diversification of metabolic gene clusters. All the data can be browsed online at www.plantcyc.org. As the authors state, these tools can be used for “systematically cataloging the chemical diversity of plants and revealing Nature’s pharmacopeia in the green world.” Plant Physiol. 10.1104/pp.16.01942 Tags: Biochemistry, Botany, Computational Biology, Genomics, Metabolism
Structure of SHR–SCR heterodimer bound to BIRD/IDD transcriptional factor JKD
 Structural biology provides a key link between genotype and phenotype. Hirano et al. probe the structure of a heterodimer of plant-specific GRAS family transcriptional regulators [SHORT-ROOT (SHR) and SCARECROW (SCR)] as bound to the transcription factor JACKDAW (JKD). The GRAS family proteins (encoded by 33 genes in Arabidopsis and 66 in rice) are involved in several signaling pathways, including root development and gibberellin and phytochrome signaling. Their analysis suggests that in GRAS proteins, the alpha-helical cap domain affects whether the proteins homo- or hetero-dimerize. The authors also found no evidence for direct DNA binding of the SHR-SCR complex, and propose that it acts as a transcription cofactors that requires complex formation with JKD for DNA binding. Nature Plants 10.1038/nplants.2017.10 Tags: Biochemistry, Development, Gene Regulation, Molecular Biology
Structural biology provides a key link between genotype and phenotype. Hirano et al. probe the structure of a heterodimer of plant-specific GRAS family transcriptional regulators [SHORT-ROOT (SHR) and SCARECROW (SCR)] as bound to the transcription factor JACKDAW (JKD). The GRAS family proteins (encoded by 33 genes in Arabidopsis and 66 in rice) are involved in several signaling pathways, including root development and gibberellin and phytochrome signaling. Their analysis suggests that in GRAS proteins, the alpha-helical cap domain affects whether the proteins homo- or hetero-dimerize. The authors also found no evidence for direct DNA binding of the SHR-SCR complex, and propose that it acts as a transcription cofactors that requires complex formation with JKD for DNA binding. Nature Plants 10.1038/nplants.2017.10 Tags: Biochemistry, Development, Gene Regulation, Molecular Biology
Phloem unloading in Arabidopsis roots
 It is well known that long distance transport and movement of molecules is enabled by phloem, but the precise mechanism of loading/unloading of phloem mobile compounds is not known. In this article, Ross-Elliott et al. used a combination of approaches (non-invasive imaging, 3D-electron microscopy, and mathematical modelling) to explore phloem unloading. The authors show that unloading of solutes in Arabidopsis roots occurs through plasmodesmata by convective unloading, a combination of mass flow and diffusion. By contrast, unloading of small proteins occurs in pulses, which they describe as batch unloading, through specialized funnel plasmodesmata, into the phloem pole pericycle. The authors also propose a new model of phloem unloading in Arabidopsis roots. (Summary by Mather Khan) eLife 10.7554/eLife.24125 Tags: Cell Biology, Physiology, Transport
It is well known that long distance transport and movement of molecules is enabled by phloem, but the precise mechanism of loading/unloading of phloem mobile compounds is not known. In this article, Ross-Elliott et al. used a combination of approaches (non-invasive imaging, 3D-electron microscopy, and mathematical modelling) to explore phloem unloading. The authors show that unloading of solutes in Arabidopsis roots occurs through plasmodesmata by convective unloading, a combination of mass flow and diffusion. By contrast, unloading of small proteins occurs in pulses, which they describe as batch unloading, through specialized funnel plasmodesmata, into the phloem pole pericycle. The authors also propose a new model of phloem unloading in Arabidopsis roots. (Summary by Mather Khan) eLife 10.7554/eLife.24125 Tags: Cell Biology, Physiology, Transport
Sterol-binding activity of PR-1 contributes to its antimicrobial activity ($)
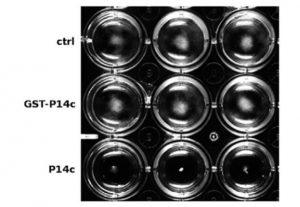 PATHOGENESIS-RELATED 1 (PR-1) protein was identified 50 years ago as a small protein induced in response to pathogens, but its mode of action has remained obscure. PR-1 is a member of the CAP family (cysteine-rich secretory protein, antigen 5, and pathogenesis-related 1). These proteins share a 150 amino acid domain that forms an α-β-α sandwich fold. Recently, yeast CAP proteins were found to bind sterols. Gamir et al. showed that PR-1 also can bind sterols, and that expression of PR-1 can complement yeast CAP mutants. Phytophthora are unable to synthesize sterols and rely on their uptake from the medium; these oomycetes are also much more sensitive to PR-1 than fungal pathogens. The authors showed that PR-1 can inhibit growth of Phytophthora brassicae in vitro, but that this inhibition is eliminated in the presence of excess cholesterol in the growth medium. PR-1 mutants blocked in sterol binding, or fusion proteins rendered too large to pass into the pathogen cell lack antimicrobial activity. Together, these studies suggest that PR-1’s mode of action is through direct removal of sterols from pathogens. Plant J. 10.1111/tpj.13398 Tags: Biochemistry, Biotic Interactions
PATHOGENESIS-RELATED 1 (PR-1) protein was identified 50 years ago as a small protein induced in response to pathogens, but its mode of action has remained obscure. PR-1 is a member of the CAP family (cysteine-rich secretory protein, antigen 5, and pathogenesis-related 1). These proteins share a 150 amino acid domain that forms an α-β-α sandwich fold. Recently, yeast CAP proteins were found to bind sterols. Gamir et al. showed that PR-1 also can bind sterols, and that expression of PR-1 can complement yeast CAP mutants. Phytophthora are unable to synthesize sterols and rely on their uptake from the medium; these oomycetes are also much more sensitive to PR-1 than fungal pathogens. The authors showed that PR-1 can inhibit growth of Phytophthora brassicae in vitro, but that this inhibition is eliminated in the presence of excess cholesterol in the growth medium. PR-1 mutants blocked in sterol binding, or fusion proteins rendered too large to pass into the pathogen cell lack antimicrobial activity. Together, these studies suggest that PR-1’s mode of action is through direct removal of sterols from pathogens. Plant J. 10.1111/tpj.13398 Tags: Biochemistry, Biotic Interactions
Proteomics of two differently pathogenic races of Fusarium oxysporum
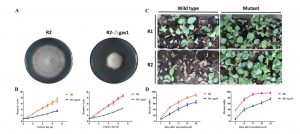 Fusarium oxysporum is a fungal pathogen of plants. F. oxysporum f. sp. conglutinans (Foc) causes fungal wilt in cabbage. Two races have been identified, with Race 2 being much more pathogenic than Race 1. Li et al. used a proteomic approach to investigate the origin of Race 2’s enhanced pathogenicity. They observed 145 proteins that differ in abundance between the two races. Proteins involved in inorganic ion transport and carbohydrate transport and metabolism were notably more abundant in the more pathogenic race, as were their corresponding mRNAs. Knocking out a gene in encoding one of these differentially expressed proteins (a glucanosyltransferase) significantly lowered the pathogenicity of both races. Sci. Reports 10.1038/srep13663 Tags: Biochemistry, Biotic Interactions
Fusarium oxysporum is a fungal pathogen of plants. F. oxysporum f. sp. conglutinans (Foc) causes fungal wilt in cabbage. Two races have been identified, with Race 2 being much more pathogenic than Race 1. Li et al. used a proteomic approach to investigate the origin of Race 2’s enhanced pathogenicity. They observed 145 proteins that differ in abundance between the two races. Proteins involved in inorganic ion transport and carbohydrate transport and metabolism were notably more abundant in the more pathogenic race, as were their corresponding mRNAs. Knocking out a gene in encoding one of these differentially expressed proteins (a glucanosyltransferase) significantly lowered the pathogenicity of both races. Sci. Reports 10.1038/srep13663 Tags: Biochemistry, Biotic Interactions
Welwitschia mirabilis sheds light on ancestral mechanisms prefiguring floral development ($)
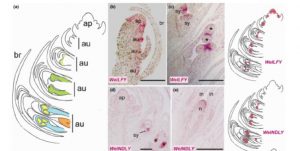 In order to look at the origins of flowers, Moyroud et al. looked at reproductive controls in the gymnosperm Welwitschia mirabilis, which, as the authors say, is a good model because, “Although the plant body is famously bizarre, the reproductive structures are generalized.” Furthermore, its reproductive structures are produced as soon as two years after germination. The authors found orthologues of key flowering genes in Welwitschia, and showed that, like it’s angiosperm counterpart, Welwitschia LFY binds to the promoter of MADS box genes corresponding to AP3/PI. Furthermore, they found that early in their development male cones showed B- and C-class MADS-box gene expression, whereas female cones showed C class expression; the same pattern as observed in angiosperms. New Phytol. 10.1111/nph.14483 Tags: Botany, Development, Molecular Biology
In order to look at the origins of flowers, Moyroud et al. looked at reproductive controls in the gymnosperm Welwitschia mirabilis, which, as the authors say, is a good model because, “Although the plant body is famously bizarre, the reproductive structures are generalized.” Furthermore, its reproductive structures are produced as soon as two years after germination. The authors found orthologues of key flowering genes in Welwitschia, and showed that, like it’s angiosperm counterpart, Welwitschia LFY binds to the promoter of MADS box genes corresponding to AP3/PI. Furthermore, they found that early in their development male cones showed B- and C-class MADS-box gene expression, whereas female cones showed C class expression; the same pattern as observed in angiosperms. New Phytol. 10.1111/nph.14483 Tags: Botany, Development, Molecular Biology
The importance of pollen chemistry in evolutionary host shifts of bees
 Some bees are generalist pollinators that gather pollen from a wide range of species, whereas others are specialists that visit only one or a few species. Vanderplanck et al. examined floral traits of the host plants of two different groups of generalist bees. There was no significant correlation between bees and the floral scent, floral reflectance, and total amino acid composition in pollen of the flowers they visit. By contrast, pollen sterol content and polypeptide content does correlate with bee preferences. This study provides insights into the evolutionary forces that affect host-shifts in bees. Sci. Reports 10.1038/srep43058 Tags: Biochemistry, Biotic Interactions, Ecophysiology
Some bees are generalist pollinators that gather pollen from a wide range of species, whereas others are specialists that visit only one or a few species. Vanderplanck et al. examined floral traits of the host plants of two different groups of generalist bees. There was no significant correlation between bees and the floral scent, floral reflectance, and total amino acid composition in pollen of the flowers they visit. By contrast, pollen sterol content and polypeptide content does correlate with bee preferences. This study provides insights into the evolutionary forces that affect host-shifts in bees. Sci. Reports 10.1038/srep43058 Tags: Biochemistry, Biotic Interactions, Ecophysiology


Leave a Reply
Want to join the discussion?Feel free to contribute!