What We’re Reading: June 21
This week we have some guest contributions from undergraduate student interns working with Professor Maria Julissa Ek-Ramos from the Universidad Autonoma de Nuevo Leon. Julissa helped the students select and read the papers, and worked with them on writing and editing the summaries, with additional editing provided by Mary Williams. What a nice learning experience. Let us know if you’d like to do something similar with your students, and enjoy.
Review: The rice genome revolution: from an ancient grain to Green Super Rice ($)
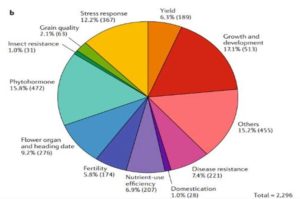 Rice is one of the major staple crops in the world as it is an essential component of diets and livelihoods. Populations in poor regions that are highly dependent on rice (Africa and South Asia) will increase dramatically by 2050, revealing the urgent need to find tools to prevent a future humanitarian crisis. Wing et al. reviewed how comparative and functional genomics of domesticated and wild rice germplasm collections provide valuable information for rice improvement. Genome mining can identify genes affecting growth and development, insect resistance, grain quality, stress response, yield, fertility, disease resistance and nutrient efficiency. The authors observe that various plant varieties have been crossed to obtain desirable phenotypes to increase the rice production. However, increasing production can also increase costs, as well as increase use of pesticides, fertilizers and water. Therefore, Green Super Rice (GSR) could be created combining characteristics of Asian rice (Oryza sativa) and African rice (Oryza glaberrima). If GSR could be produced, it could help to satisfy the demand for rice in resource-scarce regions, it might be economical in its production, and it could have the capacity to grow on marginal lands and with lower associated greenhouse gas emissions. (Summary by Gema Catalina Salas Leija and Maria Julissa Ek-Ramos). Nature Rev. Genetics 10.1038/s41576-018-0024-z.
Rice is one of the major staple crops in the world as it is an essential component of diets and livelihoods. Populations in poor regions that are highly dependent on rice (Africa and South Asia) will increase dramatically by 2050, revealing the urgent need to find tools to prevent a future humanitarian crisis. Wing et al. reviewed how comparative and functional genomics of domesticated and wild rice germplasm collections provide valuable information for rice improvement. Genome mining can identify genes affecting growth and development, insect resistance, grain quality, stress response, yield, fertility, disease resistance and nutrient efficiency. The authors observe that various plant varieties have been crossed to obtain desirable phenotypes to increase the rice production. However, increasing production can also increase costs, as well as increase use of pesticides, fertilizers and water. Therefore, Green Super Rice (GSR) could be created combining characteristics of Asian rice (Oryza sativa) and African rice (Oryza glaberrima). If GSR could be produced, it could help to satisfy the demand for rice in resource-scarce regions, it might be economical in its production, and it could have the capacity to grow on marginal lands and with lower associated greenhouse gas emissions. (Summary by Gema Catalina Salas Leija and Maria Julissa Ek-Ramos). Nature Rev. Genetics 10.1038/s41576-018-0024-z.
Review: Overview of attitudes towards genetically engineered food ($)
 The appearance and growing importance of genetically engineered (GE) food, and the extensive resistance to it, raises many issues specific to this technology. Scott et al. analyze the bases of lay opposition to GE food and evidence for how attitudes change towards this topic. The authors indicate that a lot of people are against GE food mainly because of the perceived risks and benefits, (lack of) knowledge, (lack of) trust in science, and (lack of) trust in institutions. Additionally, moral values, contagion intuitions (e.g., GE tomatoes incorporating a frost-resistance gene from the Arctic char fish nicknamed as “fish tomatoes”), political aspects and media and other resources affect their attitudes. The authors explore if any changes in attitudes toward genetic engineering can be observed by providing more information about genetic engineering and its risks and benefits. Interestingly, they found providing information does not generally increase acceptance but even can increase negative attitudes. In many cases, factual arguments are not effective, indicating a strong influence of moral or other intuitions. The authors conclude by highlighting that these reactions are based on Western developed world points of view. Taking into account that 80% of the human population lives outside of the developed world, attitudes based on their needs and opinions toward food and nature have to receive much more attention in the near future. (Summary by Jessica Carolina Seis Muñoz and Maria Julissa Ek-Ramos) Annu. Rev. Nutrition. 10.1146/annurev-nutr-071715-051223.
The appearance and growing importance of genetically engineered (GE) food, and the extensive resistance to it, raises many issues specific to this technology. Scott et al. analyze the bases of lay opposition to GE food and evidence for how attitudes change towards this topic. The authors indicate that a lot of people are against GE food mainly because of the perceived risks and benefits, (lack of) knowledge, (lack of) trust in science, and (lack of) trust in institutions. Additionally, moral values, contagion intuitions (e.g., GE tomatoes incorporating a frost-resistance gene from the Arctic char fish nicknamed as “fish tomatoes”), political aspects and media and other resources affect their attitudes. The authors explore if any changes in attitudes toward genetic engineering can be observed by providing more information about genetic engineering and its risks and benefits. Interestingly, they found providing information does not generally increase acceptance but even can increase negative attitudes. In many cases, factual arguments are not effective, indicating a strong influence of moral or other intuitions. The authors conclude by highlighting that these reactions are based on Western developed world points of view. Taking into account that 80% of the human population lives outside of the developed world, attitudes based on their needs and opinions toward food and nature have to receive much more attention in the near future. (Summary by Jessica Carolina Seis Muñoz and Maria Julissa Ek-Ramos) Annu. Rev. Nutrition. 10.1146/annurev-nutr-071715-051223.
Photochemistry beyond the red limit in chlorophyll f–containing photosystems ($)
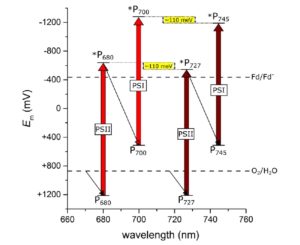 Plant scientists are familiar with the steep drop in photosynthetic activity when plants are illuminated with far-red photons (> 700 nm), because the energy of these long-wavelength photos is insufficient to initiate photochemistry at the chlorophyll a reaction center. Previously, some studies have indicated that the long-wavelength chlorophyll f can serve as an antenna complex pigment, by using heat energy to augment the energy of the longer-wavelength photons so that they can be transferred effectively to the chlorophyll a reaction center. Nürnberg et al. have now shown that chlorophyll f can in fact initiate photochemistry itself in long-wavelength light. The authors grew Chroococcidiopsis thermalis (a type of cyanobacteria) in long-wavelength light (750 nm), which led to a shift their light absorbing properties and their photosynthetic action spectra. Through studies at very low temperatures, the authors demonstrated that the chl f itself, without employing a thermal boost, can initiate photochemistry. Thus, this study provides evidence that the 700 nm photosynthetic limit is breached. (Be sure to also read the Supplementary Materials for this paper, which are excellent and provide additional analysis, interpretation and discussion). (Summary by Mary Williams) Science 10.1126/science.aar8313
Plant scientists are familiar with the steep drop in photosynthetic activity when plants are illuminated with far-red photons (> 700 nm), because the energy of these long-wavelength photos is insufficient to initiate photochemistry at the chlorophyll a reaction center. Previously, some studies have indicated that the long-wavelength chlorophyll f can serve as an antenna complex pigment, by using heat energy to augment the energy of the longer-wavelength photons so that they can be transferred effectively to the chlorophyll a reaction center. Nürnberg et al. have now shown that chlorophyll f can in fact initiate photochemistry itself in long-wavelength light. The authors grew Chroococcidiopsis thermalis (a type of cyanobacteria) in long-wavelength light (750 nm), which led to a shift their light absorbing properties and their photosynthetic action spectra. Through studies at very low temperatures, the authors demonstrated that the chl f itself, without employing a thermal boost, can initiate photochemistry. Thus, this study provides evidence that the 700 nm photosynthetic limit is breached. (Be sure to also read the Supplementary Materials for this paper, which are excellent and provide additional analysis, interpretation and discussion). (Summary by Mary Williams) Science 10.1126/science.aar8313
Novel energy transfer pathway between light-harvesting complexes and photosystem I core identified through structural studies ($)
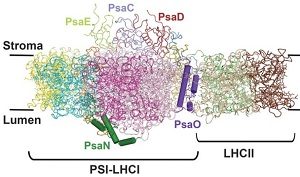 Plants must regulate the harvesting of light to maintain proper energy fluxes though photosystem I (PSI) and photosystem II (PSII). Under optimal conditions, the light-harvesting complex II (LHCII) is phosphorylated and forms a supercomplex with the PSI core and the light-harvesting complex I (LHCI). This phosphorylation is due to a kinase (STN7) that senses the redox status of the chloroplast. While there is some structural data of the interaction between PSI and PSII, it was unclear how the phosphorylation of the LHCII enhances its interaction with PSI. Pan and colleagues used single-particle cryo-electron microscopy (cryo-EM) on purified maize protein complexes to solve the PSI-LHCI-LHCII structure at a resolution of 3.3 Å. Specific subunits of PSI are important for relaying excitation to the PSI core thought chlorophyll molecules, revealing a novel pathway for energy transfer from LHCI and LCHII to the PSI core. (Summary by Julia Miller) Science 10.1126/science.aat1156
Plants must regulate the harvesting of light to maintain proper energy fluxes though photosystem I (PSI) and photosystem II (PSII). Under optimal conditions, the light-harvesting complex II (LHCII) is phosphorylated and forms a supercomplex with the PSI core and the light-harvesting complex I (LHCI). This phosphorylation is due to a kinase (STN7) that senses the redox status of the chloroplast. While there is some structural data of the interaction between PSI and PSII, it was unclear how the phosphorylation of the LHCII enhances its interaction with PSI. Pan and colleagues used single-particle cryo-electron microscopy (cryo-EM) on purified maize protein complexes to solve the PSI-LHCI-LHCII structure at a resolution of 3.3 Å. Specific subunits of PSI are important for relaying excitation to the PSI core thought chlorophyll molecules, revealing a novel pathway for energy transfer from LHCI and LCHII to the PSI core. (Summary by Julia Miller) Science 10.1126/science.aat1156
A 5-enolpyruvylshikimate 3-phosphate synthase functions as a transcriptional repressor in Populus
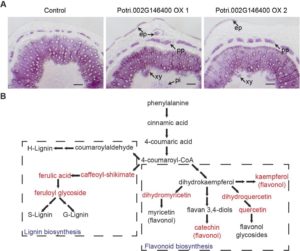 Here’s a fascinating story; starting with an association study, Xie et al. found that a protein previously identified as an enzyme involved in phenylpropanoid metabolism (specifically, 5-enolpyruvylshikimate 3-phosphate synthase, EPSP) also acts as a transcriptional regulator of this pathway, not only increasing lignin content but also the expression of dozens of other genes involved in lignin biosynthesis. Furthermore, the poplar genome has an “ordinary” EPSP gene and a second gene encoding the “dual function” enzyme. The “dual function” protein has EPSP enzymatic activity but also a DNA binding domain, encoded by an extra exon, not found in the ordinary EPSP gene. As the authors summarize, “protein moonlighting by biosynthesis enzymes, exemplified by PtrEPSP-TF,… has been proposed as an adaptive mechanism yielding novel genetic regulators in eukaryotes, where organismal complexity necessitated a complementary expansion of transcription factors.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00168
Here’s a fascinating story; starting with an association study, Xie et al. found that a protein previously identified as an enzyme involved in phenylpropanoid metabolism (specifically, 5-enolpyruvylshikimate 3-phosphate synthase, EPSP) also acts as a transcriptional regulator of this pathway, not only increasing lignin content but also the expression of dozens of other genes involved in lignin biosynthesis. Furthermore, the poplar genome has an “ordinary” EPSP gene and a second gene encoding the “dual function” enzyme. The “dual function” protein has EPSP enzymatic activity but also a DNA binding domain, encoded by an extra exon, not found in the ordinary EPSP gene. As the authors summarize, “protein moonlighting by biosynthesis enzymes, exemplified by PtrEPSP-TF,… has been proposed as an adaptive mechanism yielding novel genetic regulators in eukaryotes, where organismal complexity necessitated a complementary expansion of transcription factors.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00168
The 6xABRE synthetic promoter enables the spatiotemporal analysis of ABA-mediated transcriptional regulation
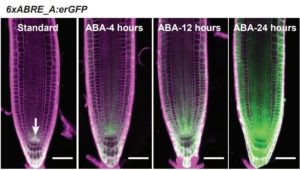 Abscisic acid (ABA) is known as stress hormone. Apart from its role in plant growth and development, it is widely studied due to its involvement in biotic and abiotic stresses. Although it had been studied for such a long time, a sensitive spatiotemporal marker for ABA is still elusive. Wu et al. developed an ABA responsive marker by using a synthetic promoter. Previous studies had shown that fusing short DNA fragments containing the ABRE (ABSCISIC ACID RESPONSIVE ELEMENT) with a minimal promoter (MP) sequence is sufficient to cause ABA-mediated expression. Here, the authors created a synthetic promoter using ABRE sequences from two well-characterized ABA responsive genes, RESPONSE TO DEHYDRATION 29A (RD29A) and ABA INSENSITIVE 1 (ABI1). The constructed marker responds to ABA in a tissue specific, time and concentration dependent manner, and is responsive to salt and mannitol. ABA-independent expression was also observed, suggesting that other, non-ABA factors interact with the synthetic promoter. Specifically, the quiescent-center specific WOX5 and NAC13 transcription factors regulate expression of the ABRE reporter. Taken together, this reporter will help to decipher stress regulation its interaction with development. (Summary by Arif Ashraf @aribidopsis) Plant Physiol. 10.1104/pp.18.00401
Abscisic acid (ABA) is known as stress hormone. Apart from its role in plant growth and development, it is widely studied due to its involvement in biotic and abiotic stresses. Although it had been studied for such a long time, a sensitive spatiotemporal marker for ABA is still elusive. Wu et al. developed an ABA responsive marker by using a synthetic promoter. Previous studies had shown that fusing short DNA fragments containing the ABRE (ABSCISIC ACID RESPONSIVE ELEMENT) with a minimal promoter (MP) sequence is sufficient to cause ABA-mediated expression. Here, the authors created a synthetic promoter using ABRE sequences from two well-characterized ABA responsive genes, RESPONSE TO DEHYDRATION 29A (RD29A) and ABA INSENSITIVE 1 (ABI1). The constructed marker responds to ABA in a tissue specific, time and concentration dependent manner, and is responsive to salt and mannitol. ABA-independent expression was also observed, suggesting that other, non-ABA factors interact with the synthetic promoter. Specifically, the quiescent-center specific WOX5 and NAC13 transcription factors regulate expression of the ABRE reporter. Taken together, this reporter will help to decipher stress regulation its interaction with development. (Summary by Arif Ashraf @aribidopsis) Plant Physiol. 10.1104/pp.18.00401
A molecular rheostat adjusts auxin flux to promote root protophloem differentiation ($)
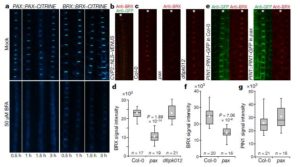 In plant development, auxin serves as a concentration-dependent signal that regulates cell differentiation, elongation and proliferation. The distribution of auxin is carried out by auxin efflux carriers such as PIN-FORMED (PIN) proteins and the specific accumulation of auxin directs organ differentiation and organization. Marhava et al. developed a mutant of the BREVIS RADIX (BRX) gene (which encodes for a plasma membrane-associated protein) and co-localizes with PIN protein at the developing protophloem sieve elements (PPSEs) in the root tip. In the brx mutants, they found that the protophloem often does not differentiate, lacks characteristic changes in the cell wall and lowers the accumulation of auxin. Furthermore, they evaluated the role of AGC kinases in the protophloem differentiation, studying an interesting protein that co-localizes with PIN at PPSEs and interacts strongly with BRX, a PROTEIN KINASE ASSOCIATED WITH BRX (PAX), by obtaining pax mutants. Results indicated pax mutants represent hypomorphic phenocopies of brx mutants, also with protophloem differentiation defects. This is interpreted as PAX and BRX act together to regulate, like a rheostat, the effects of auxin on protophloem differentiation. (Summary by Jonathan Israel Chapa García and Maria Julissa Ek-Ramos) Nature 10.1038/s41586-018-0186-z
In plant development, auxin serves as a concentration-dependent signal that regulates cell differentiation, elongation and proliferation. The distribution of auxin is carried out by auxin efflux carriers such as PIN-FORMED (PIN) proteins and the specific accumulation of auxin directs organ differentiation and organization. Marhava et al. developed a mutant of the BREVIS RADIX (BRX) gene (which encodes for a plasma membrane-associated protein) and co-localizes with PIN protein at the developing protophloem sieve elements (PPSEs) in the root tip. In the brx mutants, they found that the protophloem often does not differentiate, lacks characteristic changes in the cell wall and lowers the accumulation of auxin. Furthermore, they evaluated the role of AGC kinases in the protophloem differentiation, studying an interesting protein that co-localizes with PIN at PPSEs and interacts strongly with BRX, a PROTEIN KINASE ASSOCIATED WITH BRX (PAX), by obtaining pax mutants. Results indicated pax mutants represent hypomorphic phenocopies of brx mutants, also with protophloem differentiation defects. This is interpreted as PAX and BRX act together to regulate, like a rheostat, the effects of auxin on protophloem differentiation. (Summary by Jonathan Israel Chapa García and Maria Julissa Ek-Ramos) Nature 10.1038/s41586-018-0186-z
Feedback regulation of COOLAIR expression controls seed dormancy and flowering time ($)
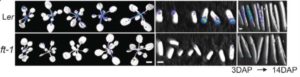 FLOWERING LOCUS C (FLC) and FLOWERING LOCUS T (FT) are proteins required for seasonal sensing in plants. Specifically, FLC and FT control seed dormancy and flowering time. In the control of flowering time, FLC expression is regulated by the prolonged-cold induction of antisense transcripts at FLC known as COOLAIR, which facilitate FLC silencing. Previously Chen et al. showed that maternal expression of FLC and FT transmits environmental information to developing seeds and therefore influence the seeds’ dormancy. Chen and Penfield studied how FT controls seed dormancy. Although FLC has been shown to act upstream of FT in the flowering time response, the authors found that in terms of seed dormancy FLC acts downstream of FT. In seed development and dormancy, warm temperatures suppress FLC expression and this response is mediated by FT. The authors conclude that, “in Arabidopsis the same genes controlled in opposite format regulate flowering time and seed dormancy in response to the temperature changes that characterize seasons.” (Summary by Verónica López Santoy and Maria Julissa Ek-Ramos) Science 10.1126/science.aar7361.
FLOWERING LOCUS C (FLC) and FLOWERING LOCUS T (FT) are proteins required for seasonal sensing in plants. Specifically, FLC and FT control seed dormancy and flowering time. In the control of flowering time, FLC expression is regulated by the prolonged-cold induction of antisense transcripts at FLC known as COOLAIR, which facilitate FLC silencing. Previously Chen et al. showed that maternal expression of FLC and FT transmits environmental information to developing seeds and therefore influence the seeds’ dormancy. Chen and Penfield studied how FT controls seed dormancy. Although FLC has been shown to act upstream of FT in the flowering time response, the authors found that in terms of seed dormancy FLC acts downstream of FT. In seed development and dormancy, warm temperatures suppress FLC expression and this response is mediated by FT. The authors conclude that, “in Arabidopsis the same genes controlled in opposite format regulate flowering time and seed dormancy in response to the temperature changes that characterize seasons.” (Summary by Verónica López Santoy and Maria Julissa Ek-Ramos) Science 10.1126/science.aar7361.
The demise of the largest and oldest African baobabs ($)
 Patrut et al. report that 8 of the 13 oldest and 5 of the 6 largest African Baobab (Adansonia digitata L.) trees, known for their enormous size and great longevity, have died (or at least their largest and/or oldest parts/stems have collapsed and died). Included in the dead are Panke, the oldest Baobab in the world at more than 2500 years, and Platland, the largest and best-known Baobab. These deaths have caused serious confusion and great concern because the cause of these significant losses is not known. Uncertainty of how, in a matter of years, something unknown can destroy what for millennia has survived puts other regions that have this type of specimens on alert. For the moment, it is not possible to determine the cause of these sudden deaths, but everything seems to point to the recent climatic changes in Africa. (Summary by Juan Alberto Mar Arriaga and Maria Julissa Ek-Ramos). Nature Plants 10.1038/s41477-018-0170-5.
Patrut et al. report that 8 of the 13 oldest and 5 of the 6 largest African Baobab (Adansonia digitata L.) trees, known for their enormous size and great longevity, have died (or at least their largest and/or oldest parts/stems have collapsed and died). Included in the dead are Panke, the oldest Baobab in the world at more than 2500 years, and Platland, the largest and best-known Baobab. These deaths have caused serious confusion and great concern because the cause of these significant losses is not known. Uncertainty of how, in a matter of years, something unknown can destroy what for millennia has survived puts other regions that have this type of specimens on alert. For the moment, it is not possible to determine the cause of these sudden deaths, but everything seems to point to the recent climatic changes in Africa. (Summary by Juan Alberto Mar Arriaga and Maria Julissa Ek-Ramos). Nature Plants 10.1038/s41477-018-0170-5.



