What We’re Reading: June 1st
Review: Beyond fossil fuel–driven nitrogen transformations ($)
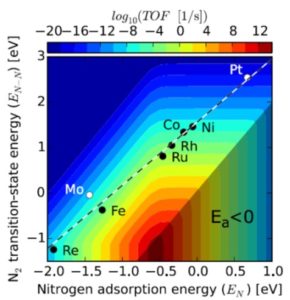 Obtaining the high yields needed to feed the human population depends on the application of nitrogen-containing fertilizers to non-leguminous crops, yet the production of these compounds consumes 1 – 2% of global energy output. Plant scientists are familiar with the conversion of N2 to NH3 from by nitrogenase (in free-living and symbiotic bacteria), as well as its industrial production via the Haber-Bosch process, but may be less familiar with other strategies being explored to produce nitrogen in forms suitable for plant food. This interesting review by Chen et al. summarizes current efforts to generate reduced nitrogen with greater energy efficiency. The article also addresses new methods to remove envorinmentally polluting nitrogen species including nitic oxide and nitrogen dioxide. Although I didn’t understand all of the details of the reactions described, the article is mostly accessible and provides a nice glimpse into the work of nitrogen chemists. (Summary by Mary Williams) Science 10.1126/science.aar6611
Obtaining the high yields needed to feed the human population depends on the application of nitrogen-containing fertilizers to non-leguminous crops, yet the production of these compounds consumes 1 – 2% of global energy output. Plant scientists are familiar with the conversion of N2 to NH3 from by nitrogenase (in free-living and symbiotic bacteria), as well as its industrial production via the Haber-Bosch process, but may be less familiar with other strategies being explored to produce nitrogen in forms suitable for plant food. This interesting review by Chen et al. summarizes current efforts to generate reduced nitrogen with greater energy efficiency. The article also addresses new methods to remove envorinmentally polluting nitrogen species including nitic oxide and nitrogen dioxide. Although I didn’t understand all of the details of the reactions described, the article is mostly accessible and provides a nice glimpse into the work of nitrogen chemists. (Summary by Mary Williams) Science 10.1126/science.aar6611
Review: Entering the next dimension – plant genomes in 3D
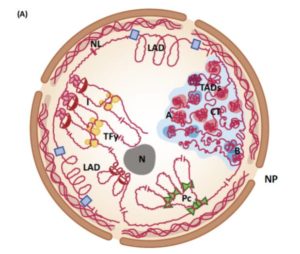 The three-dimensional arrangement of DNA within the nucleus clearly affects gene expression, but its contribution remains relatively unexplored as compared to other factors. Sotelo-Silveira et al. review new chromosome conformation capture (3C) and derived techniques for assessing genomes in 3D, as well as the corresponding biological consequences of the 3D layout. Because understanding nuclear architecture depends on knowing the sequence of the genome, our understanding is advancing rapidly thanks to cheap and fast sequencing methods, as well as methods to profile mRNA levels and epigenetic marks. As the authors report, “The latest studies also include the simultaneous characterization of chromatin accessibility, DNA methylation, histone modification, and gene expression, trying to link these characteristics to the 3D organization information.” This review provides an excellent overview of our current understanding of the 3D structure of genomes across species and cell types and during development and stress response, and highlights additional questions that remain to be resolved. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2018.03.014
The three-dimensional arrangement of DNA within the nucleus clearly affects gene expression, but its contribution remains relatively unexplored as compared to other factors. Sotelo-Silveira et al. review new chromosome conformation capture (3C) and derived techniques for assessing genomes in 3D, as well as the corresponding biological consequences of the 3D layout. Because understanding nuclear architecture depends on knowing the sequence of the genome, our understanding is advancing rapidly thanks to cheap and fast sequencing methods, as well as methods to profile mRNA levels and epigenetic marks. As the authors report, “The latest studies also include the simultaneous characterization of chromatin accessibility, DNA methylation, histone modification, and gene expression, trying to link these characteristics to the 3D organization information.” This review provides an excellent overview of our current understanding of the 3D structure of genomes across species and cell types and during development and stress response, and highlights additional questions that remain to be resolved. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2018.03.014
Chloroplast ATP synthase structure generated through cryo-EM ($)
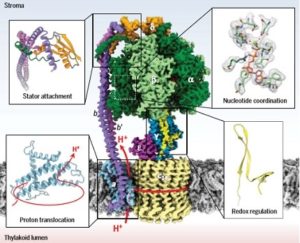 Green plants use photosynthesis to covert light into ATP (adenosine triphosphate) through the molecular action of the chloroplast F1Fo ATP synthase (cF1Fo). The ATP synthesis happens in the hydrophilic head (c F1) and is powered by the cFo rotary motor located in the photosynthetic membrane. This rotary motor is driven by the electrochemical gradient maintained across the membrane. Crystallization of this large and dynamic membrane-bound protein has been difficult so no high-resolution structure of an entire, functional ATP synthase has been generated. Thus, Hahn and colleagues used a lipid nanodisc-based purification method to generate a high-resolution spinach c F1Fo structure (2.9 Å for cF1, 3.4 Å for cFo) through cryo-electron microscopy (cryo-EM). They also characterize three functional states that are involved in rotary ATP synthesis. The features of cF1Fo are similar to ATP synthases in mitochondria and bacteria, even though the evolutionary distance is about a billion years or more. (Summary by Julia Miller) Science 10.1126/science.aat4318
Green plants use photosynthesis to covert light into ATP (adenosine triphosphate) through the molecular action of the chloroplast F1Fo ATP synthase (cF1Fo). The ATP synthesis happens in the hydrophilic head (c F1) and is powered by the cFo rotary motor located in the photosynthetic membrane. This rotary motor is driven by the electrochemical gradient maintained across the membrane. Crystallization of this large and dynamic membrane-bound protein has been difficult so no high-resolution structure of an entire, functional ATP synthase has been generated. Thus, Hahn and colleagues used a lipid nanodisc-based purification method to generate a high-resolution spinach c F1Fo structure (2.9 Å for cF1, 3.4 Å for cFo) through cryo-electron microscopy (cryo-EM). They also characterize three functional states that are involved in rotary ATP synthesis. The features of cF1Fo are similar to ATP synthases in mitochondria and bacteria, even though the evolutionary distance is about a billion years or more. (Summary by Julia Miller) Science 10.1126/science.aat4318
Response to persistent ER stress in plants: a multiphasic process that transitions cells from prosurvival activities to cell death
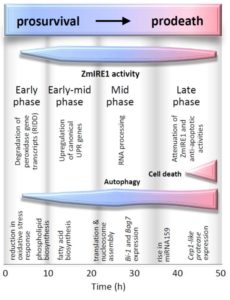 A key question in plant stress physiology is how the plant perceives stress in order to mitigate its effects. Heat (and other) stress can lead to an accumulation of unfolded or misfolded proteins in the endoplasmic reticulum, which initiates the Unfolded Protein Response (UPR), leading to a change in gene expression patterns. Srivastava et al. used several approaches to measure and track the UPR in maize seedlings, using tunicamycin as a persistent stressor. Their finding lay out a progression of activities, starting with responses that promote survival, including the selective degradation of certain mRNAs including those encoding peroxidases via Regulated IRE1-Dependent Decay (RIDD). With persistent stress, the prosurvival program is replaced by a prodeath program, which includes expression of genes including Bax inhibitor1 and genes encoding “executioner” cysteine proteases, and the induction of autophagy and cell death. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00153
A key question in plant stress physiology is how the plant perceives stress in order to mitigate its effects. Heat (and other) stress can lead to an accumulation of unfolded or misfolded proteins in the endoplasmic reticulum, which initiates the Unfolded Protein Response (UPR), leading to a change in gene expression patterns. Srivastava et al. used several approaches to measure and track the UPR in maize seedlings, using tunicamycin as a persistent stressor. Their finding lay out a progression of activities, starting with responses that promote survival, including the selective degradation of certain mRNAs including those encoding peroxidases via Regulated IRE1-Dependent Decay (RIDD). With persistent stress, the prosurvival program is replaced by a prodeath program, which includes expression of genes including Bax inhibitor1 and genes encoding “executioner” cysteine proteases, and the induction of autophagy and cell death. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00153
Phytochrome B requires PIF degradation and sequestration to induce light responses across a wide range of light conditions
 The interaction between phytochromes and phytochrome-interacting factors (PIFs) is key to light signaling. Both sequestration and degradation of PIFs occurs, and Park et al. have explored the relative contributions of these two PIF fates to the light response. They studied two mutants, one with a point mutation in the N-end (phyBG111D), and one with a C-terminal truncation accompanied by a point mutation (phyB999G767R), each retaining one of the two functions. Plants containing each of the two modified phyBs show similar responses to constant red light. The mutation that promotes PIF degradation normally but is deficient in PIF sequestration, phyBG111D, shows a more normal response to longer-scale, circadian cycles, whereas the other, which is deficient in promoting PIF degradation but sequesters PIF normally, shows a more normal response to short light pulses or sunflecks. These results suggest that PIF sequestration is more relevant to rapid, short-term light responses, whereas PIF degradation may be more important for slower, longer-scale responses. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00913
The interaction between phytochromes and phytochrome-interacting factors (PIFs) is key to light signaling. Both sequestration and degradation of PIFs occurs, and Park et al. have explored the relative contributions of these two PIF fates to the light response. They studied two mutants, one with a point mutation in the N-end (phyBG111D), and one with a C-terminal truncation accompanied by a point mutation (phyB999G767R), each retaining one of the two functions. Plants containing each of the two modified phyBs show similar responses to constant red light. The mutation that promotes PIF degradation normally but is deficient in PIF sequestration, phyBG111D, shows a more normal response to longer-scale, circadian cycles, whereas the other, which is deficient in promoting PIF degradation but sequesters PIF normally, shows a more normal response to short light pulses or sunflecks. These results suggest that PIF sequestration is more relevant to rapid, short-term light responses, whereas PIF degradation may be more important for slower, longer-scale responses. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00913
Identification of genes responsible for deceleration of circadian rhythms during tomato domestication
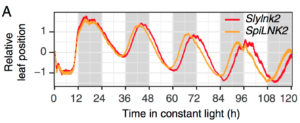 Domestication of crop plants is intimately linked to the modulation of the plants circadian rhythms, allowing adaptation to new agricultural environments. Deceleration of the clock has been instrumental in tomato domestication; however the underlying molecular mechanisms are not fully understood. In this study, Müller et al. identify a mutated allele of LNK2, together with the previously described mutation in EID1, as the main cause for the delayed circadian rhythm in cultivated tomato. Using a combination of evolutionary genomics and quantitative genetics, they show that both mutations appeared early during tomato domestication and have been fixed in the modern cultivated tomatoes. They further show that the clock deceleration by LNK2 and EID1 is light-conditional and requires a functional photoreceptor gene PHYB1. Together, this study sheds light on the molecular mechanisms associated with tomato adaptation to higher latitudes during domestication, and suggests that circadian clocks and light-signaling pathways could be engineered to obtain highly adaptable crop plants. (Summary by Matthias Benoit) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1801862115
Domestication of crop plants is intimately linked to the modulation of the plants circadian rhythms, allowing adaptation to new agricultural environments. Deceleration of the clock has been instrumental in tomato domestication; however the underlying molecular mechanisms are not fully understood. In this study, Müller et al. identify a mutated allele of LNK2, together with the previously described mutation in EID1, as the main cause for the delayed circadian rhythm in cultivated tomato. Using a combination of evolutionary genomics and quantitative genetics, they show that both mutations appeared early during tomato domestication and have been fixed in the modern cultivated tomatoes. They further show that the clock deceleration by LNK2 and EID1 is light-conditional and requires a functional photoreceptor gene PHYB1. Together, this study sheds light on the molecular mechanisms associated with tomato adaptation to higher latitudes during domestication, and suggests that circadian clocks and light-signaling pathways could be engineered to obtain highly adaptable crop plants. (Summary by Matthias Benoit) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1801862115
Decoys untangle complicated redundancy and reveal targets of circadian clock F-box proteins
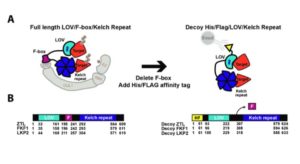 The processing of the circadian clock requires that regulatory proteins are ubiquitinated and degraded, through their interactions with F-box proteins. Lee et al. used a decoy strategy to characterize the targets of three related F-box proteins involved in clock function, ZTL, LKP2 and FKF1, which genetic studies have indicated have partially diverged, partially redundant activities. Their strategy involved replacing the F-box domains (to prevent ubiquitination of their targets) and inserting a His-tag in order to immunoprecipitate F-box protein / target complexes. Key findings include evidence for a threshold effect in the use of the decoy proteins, and the identification of distinct effects and binding partners for the three F-box proteins. The authors also demonstrate the benefits of a mammalian tissue culture cells as a heterologous ubiquitylation system. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00331
The processing of the circadian clock requires that regulatory proteins are ubiquitinated and degraded, through their interactions with F-box proteins. Lee et al. used a decoy strategy to characterize the targets of three related F-box proteins involved in clock function, ZTL, LKP2 and FKF1, which genetic studies have indicated have partially diverged, partially redundant activities. Their strategy involved replacing the F-box domains (to prevent ubiquitination of their targets) and inserting a His-tag in order to immunoprecipitate F-box protein / target complexes. Key findings include evidence for a threshold effect in the use of the decoy proteins, and the identification of distinct effects and binding partners for the three F-box proteins. The authors also demonstrate the benefits of a mammalian tissue culture cells as a heterologous ubiquitylation system. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00331
CLERK is a novel receptor kinase required for sensing of root-active CLE peptides in Arabidopsis
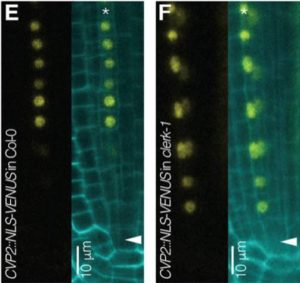 Small secreted peptides including CLEs have been identified as contributing to plant development. CLE26 and CLE45 have been shown to regulate protophloem differentiation in the root tip. Anne et al. used a combination of genetic screening and transcriptomics to identify factors downstream of these peptides. The two approaches converged on a gene encoding a previously uncharacterized leucine-rich repeat receptor kinase, which they named CLERK. They then demonstrated that CLERK is required for full sensing of root-active CLE peptides, and its topology suggests that it may act as a co-receptor for CLE peptides. CLERK is expressed in early developing protophloem, and some homologs can replace CLERK function when expressed under the CLERK promoter, suggesting that its action is context specific. (Summary by Mary Williams) Development 10.1242/dev.162354 An interview with the authors can be read here.
Small secreted peptides including CLEs have been identified as contributing to plant development. CLE26 and CLE45 have been shown to regulate protophloem differentiation in the root tip. Anne et al. used a combination of genetic screening and transcriptomics to identify factors downstream of these peptides. The two approaches converged on a gene encoding a previously uncharacterized leucine-rich repeat receptor kinase, which they named CLERK. They then demonstrated that CLERK is required for full sensing of root-active CLE peptides, and its topology suggests that it may act as a co-receptor for CLE peptides. CLERK is expressed in early developing protophloem, and some homologs can replace CLERK function when expressed under the CLERK promoter, suggesting that its action is context specific. (Summary by Mary Williams) Development 10.1242/dev.162354 An interview with the authors can be read here.
Role of BASIC PENTACYSTEIENE transcription factors in a sub-set of cytokinin signaling responses
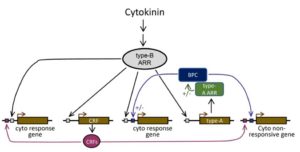 When cytokinin binds to its receptor AHKs (Arabidopsis Histidine Kinase), the signal is transduced through AHPs (Arabidopsis Histidine-containing Phosphotransfer protein) and eventually to type-B ARRs (Arabidopsis Response Regulators) transcription factors (TFs). Type-B ARR TFs mediate the primary cytokinin response, but the cytokinin response acts through other TFs in different cellular contexts. In this article, Shanks et al. studied the role of BBR/BPC (BARLEY B–RECOMBINANT/BASIC PENTACYSTEINE) TFs in cytokinin responses. The authors demonstrated that multi-BPC mutants (bpc1,2,3,4,6 and bpc1,2,3,4,6,7) have reduce cytokinin sensitivity and altered transcription of a subset of cytokinin regulated genes. Yeast two hybrid assays confirmed that BPC1 and BPC6 interact with type-B ARRs (ARR4 and ARR5), and ChIP-Seq data demonstrated that ARR10 and BPC6 bind a common promoter element in a number of genes. Furthermore, there is an overlap in genes altered in the bpc mutant with those affected in other TF mutants including CRFs (cytokinin response factors) and type-A ARRs. Altogether these data suggest that BPCs work along with known type-B ARRs in the transcriptional regulation of cytokinin responses. (Summary by Arif Ashraf @aribidopsis) Plant Journal: 10.1111/tpj.13962
When cytokinin binds to its receptor AHKs (Arabidopsis Histidine Kinase), the signal is transduced through AHPs (Arabidopsis Histidine-containing Phosphotransfer protein) and eventually to type-B ARRs (Arabidopsis Response Regulators) transcription factors (TFs). Type-B ARR TFs mediate the primary cytokinin response, but the cytokinin response acts through other TFs in different cellular contexts. In this article, Shanks et al. studied the role of BBR/BPC (BARLEY B–RECOMBINANT/BASIC PENTACYSTEINE) TFs in cytokinin responses. The authors demonstrated that multi-BPC mutants (bpc1,2,3,4,6 and bpc1,2,3,4,6,7) have reduce cytokinin sensitivity and altered transcription of a subset of cytokinin regulated genes. Yeast two hybrid assays confirmed that BPC1 and BPC6 interact with type-B ARRs (ARR4 and ARR5), and ChIP-Seq data demonstrated that ARR10 and BPC6 bind a common promoter element in a number of genes. Furthermore, there is an overlap in genes altered in the bpc mutant with those affected in other TF mutants including CRFs (cytokinin response factors) and type-A ARRs. Altogether these data suggest that BPCs work along with known type-B ARRs in the transcriptional regulation of cytokinin responses. (Summary by Arif Ashraf @aribidopsis) Plant Journal: 10.1111/tpj.13962
The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis ($)
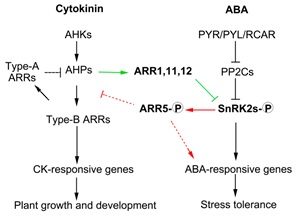 Drought stress results in a conglomeration of hormone pathways that interact to coordinate plant growth and stress response. ABA and cytokinin signaling are known to be antagonistic but it is unclear how these two pathways are connected. Huang et al. identified roles for several ARR proteins, negative regulators in cytokinin signaling, in the ABA pathway. Specifically, ABA promoting kinases, SnRK2s, were found to phosphorylate ARR5, promoting ARR5 stability and enhancing cytokinin inhibition and enhancing ABA signaling. Meanwhile, ARR1,11,12 inhibit SnRK2s to downregulate ABA signaling. These data show a model in which there is crosstalk between ABA and cytokinin signaling components. (Summary by Alecia Biel) Mol. Plant 10.1016/j.molp.2018.05.001.
Drought stress results in a conglomeration of hormone pathways that interact to coordinate plant growth and stress response. ABA and cytokinin signaling are known to be antagonistic but it is unclear how these two pathways are connected. Huang et al. identified roles for several ARR proteins, negative regulators in cytokinin signaling, in the ABA pathway. Specifically, ABA promoting kinases, SnRK2s, were found to phosphorylate ARR5, promoting ARR5 stability and enhancing cytokinin inhibition and enhancing ABA signaling. Meanwhile, ARR1,11,12 inhibit SnRK2s to downregulate ABA signaling. These data show a model in which there is crosstalk between ABA and cytokinin signaling components. (Summary by Alecia Biel) Mol. Plant 10.1016/j.molp.2018.05.001.
Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis
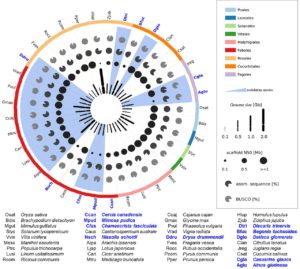 The ability to participate in nitrogen-fixing root nodule symbiosis shows an interesting phylogenetic pattern, with some families showing a large number of nodulating species interspersed with non-nodulating ones, and some families showing only a few nodulating species. A current model suggests that in some families, there is a pre-disposition towards nodulation, which lowers the bar for the full acquisition of this trait. To further test this model, Griesmann et al. have assembled genome sequences for many additional nodulating and non-nodulating species. Interestingly, their data point to a different model, in which the nodulation trait has been selectively lost, specifically through loss-of-function of one of two genes, NIN and RPG. Critically, their study suggests a stronger-than expected negative selection against symbiosis, which needs to be considered in efforts to engineer nitrogen-fixing symbiosis into crop plants. (Summary by Mary Williams) Science 10.1126/science.aat1743
The ability to participate in nitrogen-fixing root nodule symbiosis shows an interesting phylogenetic pattern, with some families showing a large number of nodulating species interspersed with non-nodulating ones, and some families showing only a few nodulating species. A current model suggests that in some families, there is a pre-disposition towards nodulation, which lowers the bar for the full acquisition of this trait. To further test this model, Griesmann et al. have assembled genome sequences for many additional nodulating and non-nodulating species. Interestingly, their data point to a different model, in which the nodulation trait has been selectively lost, specifically through loss-of-function of one of two genes, NIN and RPG. Critically, their study suggests a stronger-than expected negative selection against symbiosis, which needs to be considered in efforts to engineer nitrogen-fixing symbiosis into crop plants. (Summary by Mary Williams) Science 10.1126/science.aat1743
NLR mutations suppressing immune hybrid incompatibility and their effects on disease resistance
 Hybrid incompatibility (HI) is effectively the opposite of hybrid vigor, and indicates a deleterious effect arising from the combination of two genomes. Because of reduced fitness of the hybrids, HI can be considered as contributing towards speciation. In Arabidopsis thaliana, HI has been recognized in crosses between some accessions including between Landsberg (Ler) and Kashmir-2 (Kas-2). The hybrids show enhanced levels of defense gene expression, and disease resistance genes (R genes) have previously been implicated as contributing to HI. Atanasov et al. used a genetic approach to identify suppressors of Ler/Kas-2 incompatibility (sulki) mutants, which they mapped to a cluster of R genes. The authors speculate that detrimental autoimmune phenotype could arise due to interactions between proteins encoded by the two parental loci. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00462
Hybrid incompatibility (HI) is effectively the opposite of hybrid vigor, and indicates a deleterious effect arising from the combination of two genomes. Because of reduced fitness of the hybrids, HI can be considered as contributing towards speciation. In Arabidopsis thaliana, HI has been recognized in crosses between some accessions including between Landsberg (Ler) and Kashmir-2 (Kas-2). The hybrids show enhanced levels of defense gene expression, and disease resistance genes (R genes) have previously been implicated as contributing to HI. Atanasov et al. used a genetic approach to identify suppressors of Ler/Kas-2 incompatibility (sulki) mutants, which they mapped to a cluster of R genes. The authors speculate that detrimental autoimmune phenotype could arise due to interactions between proteins encoded by the two parental loci. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00462