What We’re Reading: July 20th
This week we have a short edition as the editor and many of the contributors have been busy at the Plant Biology meeting in Montréal, Canada. This was a hugely successful conference by all accounts. You can get a glimpse of it from the level of activity on Twitter – see #PlantBio18. Besides great scientific talks, the meeting provided opportunities for workshops on topics from reproducibility to science communication, and informal chats about careers, ASPB governance and more. It was also a chance to meet up with the Plantae Fellows (many of whom contribute regularly to this series) and the Conviron Scholars (and to announce the next Conviron Scholars program – please share with interested students, applications due by August 13). A “town hall” meeting provided a chance for members of the community to raise issues they’d like to see addressed and showed the need for ongoing conversation outside of the meeting boundaries. Watch this space for more information about these conversations.
And mark your calendars – next year we meet at #PlantBio19 in San Jose, California, August 3 – 7th. I’ll be there!
Review: Applying synthetic biology and genetic engineering to photosynthesis
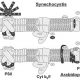
 Photosynthesis is a complex process that has the potential to be greatly improved through human modifications. However, these modifications have been limited by two main factors: the high degree of conservation of the components, and the multiprotein complexes that require specifically modified proteins from different cellular compartments. Nevertheless, the light reactions of photosynthesis are exciting targets for genetic engineering and synthetic biology due to their importance in photosynthesis and their further uses in natural compound and energy production. In this review, Dario Leister describes current advances in modifying the light reactions of photosynthesis and what future work is required. He also discusses how modified photosynthetic processes can be used to produce hydrogen, electricity or other high-value compounds. (Summary by Julia Miller) Plant Physiol. 10.1104/pp.18.00360
Photosynthesis is a complex process that has the potential to be greatly improved through human modifications. However, these modifications have been limited by two main factors: the high degree of conservation of the components, and the multiprotein complexes that require specifically modified proteins from different cellular compartments. Nevertheless, the light reactions of photosynthesis are exciting targets for genetic engineering and synthetic biology due to their importance in photosynthesis and their further uses in natural compound and energy production. In this review, Dario Leister describes current advances in modifying the light reactions of photosynthesis and what future work is required. He also discusses how modified photosynthetic processes can be used to produce hydrogen, electricity or other high-value compounds. (Summary by Julia Miller) Plant Physiol. 10.1104/pp.18.00360
Co-expression networks for strawberry flower and fruit development
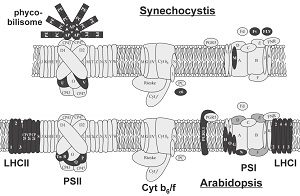
 The diploid strawberry (Fragaria vesca) is a useful model system for understanding non-climacteric ripening and seed-to-fruit cross-tissues communication. Previously, spatial and temporal transcriptome data from the strawberry fruit and flowers was generated. Shahan and colleagues combine the transcriptome data with new RNA-seq libraries to create co-expression networks. They used post-hoc consensus clustering approach and bootstrapping to generate reproducible and robust clusters from this data set. Based on the networks, they found increased iron transport from the receptacle to the seed after fertilization, and characterized a floral mutant and the underlying gene. These networks are available on the web interface www.fv.rosaceaefruits.org. The networks can be used to further mine the transcriptome data, and can be used to build a framework for understanding other species in the Rosaceae. (Summary by Julia Miller) Plant Physiol. 10.1104/pp.18.00086
The diploid strawberry (Fragaria vesca) is a useful model system for understanding non-climacteric ripening and seed-to-fruit cross-tissues communication. Previously, spatial and temporal transcriptome data from the strawberry fruit and flowers was generated. Shahan and colleagues combine the transcriptome data with new RNA-seq libraries to create co-expression networks. They used post-hoc consensus clustering approach and bootstrapping to generate reproducible and robust clusters from this data set. Based on the networks, they found increased iron transport from the receptacle to the seed after fertilization, and characterized a floral mutant and the underlying gene. These networks are available on the web interface www.fv.rosaceaefruits.org. The networks can be used to further mine the transcriptome data, and can be used to build a framework for understanding other species in the Rosaceae. (Summary by Julia Miller) Plant Physiol. 10.1104/pp.18.00086
Auxin regulates shoot stem cells
 Auxin and cytokinin play critical roles in determination of cell fate in the shoot apical meristem (SAM). Until now auxin’s role was thought to be confined to the peripheral zone (PZ), promoting cell differentiation and organogenesis, mediated by AR5/MP. However, studies using GC-MS have shown the presence of IAA in the central zone (CZ). MP also has been found to be present not only in the PZ but also in the CZ. In order to elucidate the possible function of MP in auxin signaling in stem cells, Luo and co-workers crossed the mp mutant with the stem cell overproliferating clv3. With this double mutant they were able to have enough stem cells to identify possible targets of MP. DRN was identified as a target and negatively regulated by MP. They also demonstrated that DRN positively regulates CLV3. The authors concluded that MP-DRN mediates negative auxin regulation of CLV3 to regulate the stem cell pool. (Summary by Cecilia Vasquez-Robinet) Mol Plant 10.1016/j.molp.2018.04.006
Auxin and cytokinin play critical roles in determination of cell fate in the shoot apical meristem (SAM). Until now auxin’s role was thought to be confined to the peripheral zone (PZ), promoting cell differentiation and organogenesis, mediated by AR5/MP. However, studies using GC-MS have shown the presence of IAA in the central zone (CZ). MP also has been found to be present not only in the PZ but also in the CZ. In order to elucidate the possible function of MP in auxin signaling in stem cells, Luo and co-workers crossed the mp mutant with the stem cell overproliferating clv3. With this double mutant they were able to have enough stem cells to identify possible targets of MP. DRN was identified as a target and negatively regulated by MP. They also demonstrated that DRN positively regulates CLV3. The authors concluded that MP-DRN mediates negative auxin regulation of CLV3 to regulate the stem cell pool. (Summary by Cecilia Vasquez-Robinet) Mol Plant 10.1016/j.molp.2018.04.006
Arabidopsis thaliana-associated Pseudomonas diversity and evolution
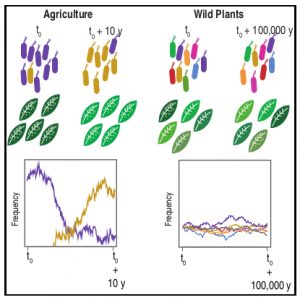 There is not much available information regarding the evolutionary aspects of one of the most studied pathosystem in plant biology: Arabidopsis thaliana and Pseudomonas. Karazov et al. performed various surveys in Arabidopsis wild populations, for the detection and characterization of their associated Pseudomonas strains. Pseudomonas spp. were detected in 97% of epi- and 88% of endophytic samples, and classified into 56 operational taxonomic units (OTUs) according to 16S sequence similarity. OTU5, classified as P. viridiflava, was found in 59% of epi- and 58% of endophytic samples, making it the predominant lineage within the isolates. Whole genome sequence of the OTU5 isolates led to the classification of 1355 isolates into 82 different strains, closely related to P. viridiflava and P. syringae. Five from the OTU5 strains accounted for 46% of the sequenced isolates, while non-OTU5 strains were rarely overrepresented in several isolates. 26 of the OTU5 strains were tested for their pathogenicity in the Eyach 15-2 Arabidopsis genotype; all but one of the assessed strains reduced plant growth, and some of these strains were found to be even more virulent than the highly virulent Arabidopsis pathogen P. syringae pv. tomato DC3000. Finally, through phylogenetic analyses, the authors were able to predict that strains belonging to OTU5 diverged approximately 300,000 years ago, maintaining nonetheless their ability to successfully colonize Arabidopsis, and contributing to a high microbial diversity in wild populations of this plant. (Summary by Juan S. Ramirez-Prado) Cell Host Microb. 10.1016/j.chom.2018.06.011
There is not much available information regarding the evolutionary aspects of one of the most studied pathosystem in plant biology: Arabidopsis thaliana and Pseudomonas. Karazov et al. performed various surveys in Arabidopsis wild populations, for the detection and characterization of their associated Pseudomonas strains. Pseudomonas spp. were detected in 97% of epi- and 88% of endophytic samples, and classified into 56 operational taxonomic units (OTUs) according to 16S sequence similarity. OTU5, classified as P. viridiflava, was found in 59% of epi- and 58% of endophytic samples, making it the predominant lineage within the isolates. Whole genome sequence of the OTU5 isolates led to the classification of 1355 isolates into 82 different strains, closely related to P. viridiflava and P. syringae. Five from the OTU5 strains accounted for 46% of the sequenced isolates, while non-OTU5 strains were rarely overrepresented in several isolates. 26 of the OTU5 strains were tested for their pathogenicity in the Eyach 15-2 Arabidopsis genotype; all but one of the assessed strains reduced plant growth, and some of these strains were found to be even more virulent than the highly virulent Arabidopsis pathogen P. syringae pv. tomato DC3000. Finally, through phylogenetic analyses, the authors were able to predict that strains belonging to OTU5 diverged approximately 300,000 years ago, maintaining nonetheless their ability to successfully colonize Arabidopsis, and contributing to a high microbial diversity in wild populations of this plant. (Summary by Juan S. Ramirez-Prado) Cell Host Microb. 10.1016/j.chom.2018.06.011