What We’re Reading: January 18th
Physiologia Plantarum Special Issue: Root Biology
 The year 2019 kicks off with a special issue on root biology, with all articles free to access for six months. Topics include interactions of roots with parasites and symbionts, root branching, transport in the root system, and roots of woody species. (Summary by Mary Williams) Physiologia Plantarum January 2019
The year 2019 kicks off with a special issue on root biology, with all articles free to access for six months. Topics include interactions of roots with parasites and symbionts, root branching, transport in the root system, and roots of woody species. (Summary by Mary Williams) Physiologia Plantarum January 2019
Review: Computational approaches to design and test plant synthetic metabolic pathways
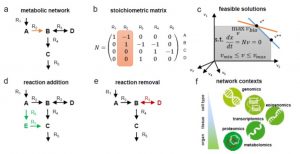 As photosynthetic organisms, plants are desirable and potentially cost-effective chassis for the production of novel compounds, but they are also inherently metabolically complex. Synthetic metabolic pathways can also improve upon nature by making plants more metabolically efficient.
As photosynthetic organisms, plants are desirable and potentially cost-effective chassis for the production of novel compounds, but they are also inherently metabolically complex. Synthetic metabolic pathways can also improve upon nature by making plants more metabolically efficient. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field ($)
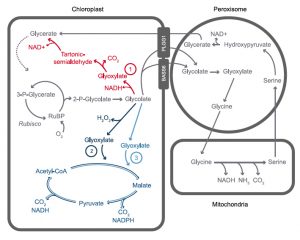 Rubisco uses CO2 from the air to carboxylate its substrate RuBP, resulting in an increase in fixed carbon. However, rubisco can also oxygenate RuBP and this leads to the production of glycolate. Plants must invest energy in recycling this toxic by-product in a process called photorespiration, resulting in a loss of fixed carbon. South et al attempted to improve photosynthesis and boost yields by recycling glycolate through more efficient pathways. They engineered three photorespiratory bypasses independently into tobacco, and performed glasshouse and field trials on the transgenic plants. All three pathways increased total biomass to varying degrees. The pathway conferring the biggest gain in biomass consisted of an algal glycolate dehydrogenase and a malate synthase from squash. This bypass showed a further enhancement in biomass (40% higher than wild type) when coupled with RNAi suppression of a chloroplast glycolate transporter (PLGG1), which reduced the capacity of glycolate to enter the native photorespiratory pathway. The authors also observed that these plants had an improved photosynthetic rate, and concluded that this was a result of CO2 release inside the chloroplast as a consequence of the introduced pathway. Having demonstrated the power of this approach in tobacco, engineering these pathways into crop species is now hotly anticipated. (Summary by Mike Page) Science 10.1126/science.aat9077
Rubisco uses CO2 from the air to carboxylate its substrate RuBP, resulting in an increase in fixed carbon. However, rubisco can also oxygenate RuBP and this leads to the production of glycolate. Plants must invest energy in recycling this toxic by-product in a process called photorespiration, resulting in a loss of fixed carbon. South et al attempted to improve photosynthesis and boost yields by recycling glycolate through more efficient pathways. They engineered three photorespiratory bypasses independently into tobacco, and performed glasshouse and field trials on the transgenic plants. All three pathways increased total biomass to varying degrees. The pathway conferring the biggest gain in biomass consisted of an algal glycolate dehydrogenase and a malate synthase from squash. This bypass showed a further enhancement in biomass (40% higher than wild type) when coupled with RNAi suppression of a chloroplast glycolate transporter (PLGG1), which reduced the capacity of glycolate to enter the native photorespiratory pathway. The authors also observed that these plants had an improved photosynthetic rate, and concluded that this was a result of CO2 release inside the chloroplast as a consequence of the introduced pathway. Having demonstrated the power of this approach in tobacco, engineering these pathways into crop species is now hotly anticipated. (Summary by Mike Page) Science 10.1126/science.aat9077
Genome-scale fluxome of Synechococcus elongatus UTEX 2973 using transient 13C-labeling data
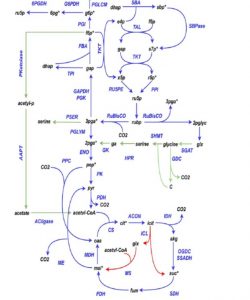 Synechococcus elongates UTEX 2973 (also known as Synechococcus 2973) has the fastest doubling time of known cyanobacteria, completely replicating itself in just over two hours. This fast growth rate makes it an interesting platform for industrial applications. Hendry et al set out to understand what makes it so fast, using genome-scale Isotopic Non-Stationary-13C-Metabolic Flux Analysis (INST-MFA). They found that the carbon-metabolism of Synechococcus 2973 deviates in several ways from that of the model species Synechocystis 6803, including a higher rate of CO2 assimilation arising from elevated rates of ATP and NADPH generation, and the extensive reassimilation of CO2 liberated by photorespiration; together these lead to the astonishing conversion of >96% of assimilated CO2 into biomass. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.01357
Synechococcus elongates UTEX 2973 (also known as Synechococcus 2973) has the fastest doubling time of known cyanobacteria, completely replicating itself in just over two hours. This fast growth rate makes it an interesting platform for industrial applications. Hendry et al set out to understand what makes it so fast, using genome-scale Isotopic Non-Stationary-13C-Metabolic Flux Analysis (INST-MFA). They found that the carbon-metabolism of Synechococcus 2973 deviates in several ways from that of the model species Synechocystis 6803, including a higher rate of CO2 assimilation arising from elevated rates of ATP and NADPH generation, and the extensive reassimilation of CO2 liberated by photorespiration; together these lead to the astonishing conversion of >96% of assimilated CO2 into biomass. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.01357
A cis‐acting bidirectional transcription switch controls sexual dimorphism in the liverwort
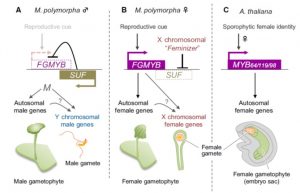 Bryophytes spend most of their lifecycle in the haploid, gametophytic form, of which there are two types, male (sperm forming) and female (egg forming). Hisanaga et al. investigated the genetic basis that determines sex in the model liverwort Marchantia polymorpha. Their findings are fascinating. A single genetic locus acts as a switch, to specify either male or female form. MpFGMYB encodes a transcription factor specifically expressed in females; loss-of-function alleles cause a female-to-male conversion. In males, the opposite DNA strand is transcribed, producing a long non-coding RNA that silences MpFGMYB. This system thus acts as a toggle, switching the organism into either the male or female state. (Summary by Mary Williams) EMBO J. 10.15252/embj.2018100240
Bryophytes spend most of their lifecycle in the haploid, gametophytic form, of which there are two types, male (sperm forming) and female (egg forming). Hisanaga et al. investigated the genetic basis that determines sex in the model liverwort Marchantia polymorpha. Their findings are fascinating. A single genetic locus acts as a switch, to specify either male or female form. MpFGMYB encodes a transcription factor specifically expressed in females; loss-of-function alleles cause a female-to-male conversion. In males, the opposite DNA strand is transcribed, producing a long non-coding RNA that silences MpFGMYB. This system thus acts as a toggle, switching the organism into either the male or female state. (Summary by Mary Williams) EMBO J. 10.15252/embj.2018100240
The APETALA2-like transcription factor SUPERNUMERARY BRACT controls rice seed shattering and seed size
 Non-shattering seed, or seed that stays attached to the stem when mature, is a key domestication trait that makes harvesting easier. Previous studies have identified several domestication genes that suppress the formation of the seed’s abscission zone, thus preventing shattering. Jiang et al. used a genetic approach to identify ssh1, which suppresses seed shattering; SSH1 encodes the AP2-type transcription factor SUPERNUMERARY BRACT (SNB). The development of the absission zone is positively correlated with SNB expression. Lignin deposition at the abscission zone is increased in the ssh1 mutant, and two genes previously identified as involved in seed shattering are downregulated in the ssh1 mutant; SNB likely is a direct transcriptional activator of these genes. Interestingly, the ssh1 mutant also has increased seed size and weight, which may arise due to other transcriptional targets, possibly involving hormones. This study provide new targets for engineering increased grain yields. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00304
Non-shattering seed, or seed that stays attached to the stem when mature, is a key domestication trait that makes harvesting easier. Previous studies have identified several domestication genes that suppress the formation of the seed’s abscission zone, thus preventing shattering. Jiang et al. used a genetic approach to identify ssh1, which suppresses seed shattering; SSH1 encodes the AP2-type transcription factor SUPERNUMERARY BRACT (SNB). The development of the absission zone is positively correlated with SNB expression. Lignin deposition at the abscission zone is increased in the ssh1 mutant, and two genes previously identified as involved in seed shattering are downregulated in the ssh1 mutant; SNB likely is a direct transcriptional activator of these genes. Interestingly, the ssh1 mutant also has increased seed size and weight, which may arise due to other transcriptional targets, possibly involving hormones. This study provide new targets for engineering increased grain yields. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00304
A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate
 Plants respond to a low phosphate environment through increased elongation of root hairs. Bhosale et al. showed that low external P increases levels of the auxin (IAA) in Arabidopsis roots through the TAA1 (TRYPTOPHAN AMINO TRANSFERASE 1)-mediated auxin biosynthesis pathway. At the same time, the auxin influx carrier AUX1 facilitates shootward auxin transport in low-P stress condition; expressing AUX1 in lateral root cap (LRC) and epidermis is sufficient to complement the root hair phenotype of aux1 under low P stress. Increased IAA induces multiple transcription factors in a spatial manner such as ARF19 in the root apex, RSL2 (ROOT HAIR DEFECTIVE 6-LIKE 2) and RSL4 in the elongation and differentiation zones, respectively, to promote root hair elongation. This work provided a comprehensive molecular mechanism of root hair elongation under low P conditions. (Summary by Arif Ashraf) Nature Communications: 10.1038/s41467-018-03851-3
Plants respond to a low phosphate environment through increased elongation of root hairs. Bhosale et al. showed that low external P increases levels of the auxin (IAA) in Arabidopsis roots through the TAA1 (TRYPTOPHAN AMINO TRANSFERASE 1)-mediated auxin biosynthesis pathway. At the same time, the auxin influx carrier AUX1 facilitates shootward auxin transport in low-P stress condition; expressing AUX1 in lateral root cap (LRC) and epidermis is sufficient to complement the root hair phenotype of aux1 under low P stress. Increased IAA induces multiple transcription factors in a spatial manner such as ARF19 in the root apex, RSL2 (ROOT HAIR DEFECTIVE 6-LIKE 2) and RSL4 in the elongation and differentiation zones, respectively, to promote root hair elongation. This work provided a comprehensive molecular mechanism of root hair elongation under low P conditions. (Summary by Arif Ashraf) Nature Communications: 10.1038/s41467-018-03851-3
Cytokinin modulates context-dependent chromatin accessibility through the type-B response regulators ($)
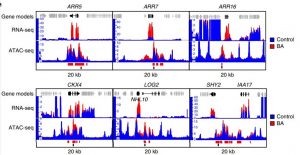 The cytokinin-based response is assumed to be achieved through context-dependent transcriptional regulatory mechanisms. Potter et al. analyzed cytokinin-induced alterations in chromatin accessibility using ATAC-seq (Assay for Transposase-Accessible Chromatin using Sequencing) and combined these results with RNA-seq and chromatin immunoprecipitation (ChIP)-seq datasets. Their results suggest that differentially accessible regions are adjacent to cytokinin-regulated genes and mostly overlap with type-B ARR ChIP peaks. This article illustrates the dynamics between cytokinin and chromatin to facilitate tissue-specific transcriptional regulation for plant development and stress response. (Summary by Arif Ashraf) Nature Plants 10.1038/s4147-018-0290-y
The cytokinin-based response is assumed to be achieved through context-dependent transcriptional regulatory mechanisms. Potter et al. analyzed cytokinin-induced alterations in chromatin accessibility using ATAC-seq (Assay for Transposase-Accessible Chromatin using Sequencing) and combined these results with RNA-seq and chromatin immunoprecipitation (ChIP)-seq datasets. Their results suggest that differentially accessible regions are adjacent to cytokinin-regulated genes and mostly overlap with type-B ARR ChIP peaks. This article illustrates the dynamics between cytokinin and chromatin to facilitate tissue-specific transcriptional regulation for plant development and stress response. (Summary by Arif Ashraf) Nature Plants 10.1038/s4147-018-0290-y
Plant PRC2 subunit VRN2 stability regulation through oxygen dependent proteolysis
 Polycomb repressive complex 2 (PRC2) represses the expression of its gene targets epigenetically and VERNALIZATION 2 (VRN2) is a subunit of the PRC2 complex in Arabidopsis. The PRC2-VRN2 complex, among other roles, regulates flowering after long cold exposure. Gibbs and coworkers identified VRN2 as a possible target for the N end rule pathway, since it posses a conserved Met-Cys (MC-) N terminal. The N-end rule regulates protein destruction by ubiquitylation with specific E3 ligases of the N terminal (N-) degron. Using the PRT6 E3-ligase mutant and an Ala-VRN2 mutated version they could show that the N-end rule pathway indeed regulates VRN2 under non stressing conditions, keeping it only in meristematic tissues. Further studies also showed that VRN2 is stabilized through the plant by low levels of O2 and contributes to hypoxia resistance. Experiments with the VRN2 barley homolog also showed a functional N-degron. Phylogenetic analysis of other VRN2 plant homologs suggested that this mechanism appeared early in angiosperm evolution, linking PRC2 epigenetic activity to environmental conditions. (Summary by Cecilia Vasquez Robinet) Nature Comms. 10.1038/s41467-018-07875-7
Polycomb repressive complex 2 (PRC2) represses the expression of its gene targets epigenetically and VERNALIZATION 2 (VRN2) is a subunit of the PRC2 complex in Arabidopsis. The PRC2-VRN2 complex, among other roles, regulates flowering after long cold exposure. Gibbs and coworkers identified VRN2 as a possible target for the N end rule pathway, since it posses a conserved Met-Cys (MC-) N terminal. The N-end rule regulates protein destruction by ubiquitylation with specific E3 ligases of the N terminal (N-) degron. Using the PRT6 E3-ligase mutant and an Ala-VRN2 mutated version they could show that the N-end rule pathway indeed regulates VRN2 under non stressing conditions, keeping it only in meristematic tissues. Further studies also showed that VRN2 is stabilized through the plant by low levels of O2 and contributes to hypoxia resistance. Experiments with the VRN2 barley homolog also showed a functional N-degron. Phylogenetic analysis of other VRN2 plant homologs suggested that this mechanism appeared early in angiosperm evolution, linking PRC2 epigenetic activity to environmental conditions. (Summary by Cecilia Vasquez Robinet) Nature Comms. 10.1038/s41467-018-07875-7
Chloroplasts navigate towards the pathogen interface to counteract Phytophthora infection
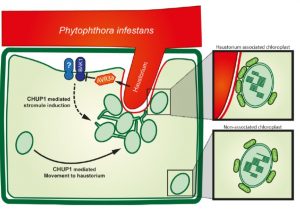 Chloroplasts have diverse roles in plant defense, including contributing to the production of defense compounds. Toufexi, Duggan et al. show new data indicating the dynamic relocation of chloroplasts to the contact point of the oomycete pathogen Phytophthora infestans. Infection also causes an increase of stromule formation (stromules are chloroplast membrane extensions implicated in defense). The authors also demonstrated a role for the chloroplast outer-membrane protein CHUP1 in chloroplast recruitment to the pathogen interface and stromule induction. Signaling via BAK1, targeted by the funal effector AVR3a, contributes to these defense responses. Be sure to have a look at the nice movies! (Summary by Mary Williams) bioRxiv 10.1101/516443
Chloroplasts have diverse roles in plant defense, including contributing to the production of defense compounds. Toufexi, Duggan et al. show new data indicating the dynamic relocation of chloroplasts to the contact point of the oomycete pathogen Phytophthora infestans. Infection also causes an increase of stromule formation (stromules are chloroplast membrane extensions implicated in defense). The authors also demonstrated a role for the chloroplast outer-membrane protein CHUP1 in chloroplast recruitment to the pathogen interface and stromule induction. Signaling via BAK1, targeted by the funal effector AVR3a, contributes to these defense responses. Be sure to have a look at the nice movies! (Summary by Mary Williams) bioRxiv 10.1101/516443
Root branching toward water involves posttranslational modification of transcription factor ARF7 ($)
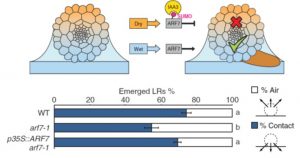 Roots do amazing things, particularly in terms of how they optimize their growth in response to their very local environment including water, nutrients, microbes, and physical obstructions. Branching in regions of contact with water is called hydropatterning, and previous studies showed that an auxin gradient forms in response to differential water contact. Orosa-Puente, Leftley, von Wangenheim et al. now show that the transcription factor ARF7 is also involved, and its action is mediated in part by one of its transcriptional targets, LBD16. Their study finds that on the wet side of the root, ARF7 is active and promotes lateral root formation, whereas on the dry side ARF7 is posttranslationally modified through SUMOylation, promoting interaction with the IAA3 repressor protein. (Summary by Mary Williams) Science 10.1126/science.aau3956
Roots do amazing things, particularly in terms of how they optimize their growth in response to their very local environment including water, nutrients, microbes, and physical obstructions. Branching in regions of contact with water is called hydropatterning, and previous studies showed that an auxin gradient forms in response to differential water contact. Orosa-Puente, Leftley, von Wangenheim et al. now show that the transcription factor ARF7 is also involved, and its action is mediated in part by one of its transcriptional targets, LBD16. Their study finds that on the wet side of the root, ARF7 is active and promotes lateral root formation, whereas on the dry side ARF7 is posttranslationally modified through SUMOylation, promoting interaction with the IAA3 repressor protein. (Summary by Mary Williams) Science 10.1126/science.aau3956



