Using a historic wheat collection to revolutionize breeding
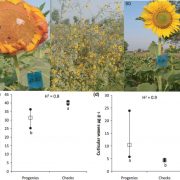
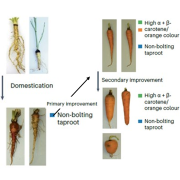
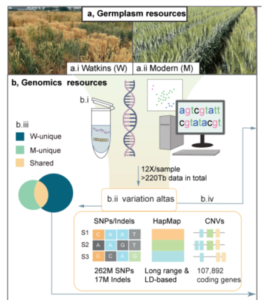 Wheat (Triticum aestivum) is an important crop, making up ~20% of food consumed worldwide. As the population grows, wheat yields must increase to meet demand. To achieve this, genetically diverse wheat landraces (aka traditional varieties, which are locally adapted cultivars) can be utilized. Here, Cheng et al. whole-genome sequenced 827 wheat landraces from the Watkins collection, which was collected in the 1920’s and 1930’s from 32 countries. These were divided into seven groups, five of which do not overlap with modern wheat, hence containing novel diversity. Further analysis identified 162 million single nucleotide polymorphisms (SNPs) in the Watkins collection which are absent from modern wheat. Nearly 14000 of these affect gene function and are in genes that have no variation in elite wheat cultivars. Ontology term analysis revealed that these genes are associated with yield, disease resistance and stress tolerance, so these SNPs could be important for breeding. The authors introduced some of this variation into the modern wheat Paragon and used field trials to demonstrate how the Watkins collection could be used in pre-breeding. Overall, this study provides a plethora of data that could revolutionize breeding; to do so the authors have made the data available at https://wwwg2b.com/. (Summary by Rose McNelly @Rose_McN) Nature 10.1038/s41586-024-07682-9
Wheat (Triticum aestivum) is an important crop, making up ~20% of food consumed worldwide. As the population grows, wheat yields must increase to meet demand. To achieve this, genetically diverse wheat landraces (aka traditional varieties, which are locally adapted cultivars) can be utilized. Here, Cheng et al. whole-genome sequenced 827 wheat landraces from the Watkins collection, which was collected in the 1920’s and 1930’s from 32 countries. These were divided into seven groups, five of which do not overlap with modern wheat, hence containing novel diversity. Further analysis identified 162 million single nucleotide polymorphisms (SNPs) in the Watkins collection which are absent from modern wheat. Nearly 14000 of these affect gene function and are in genes that have no variation in elite wheat cultivars. Ontology term analysis revealed that these genes are associated with yield, disease resistance and stress tolerance, so these SNPs could be important for breeding. The authors introduced some of this variation into the modern wheat Paragon and used field trials to demonstrate how the Watkins collection could be used in pre-breeding. Overall, this study provides a plethora of data that could revolutionize breeding; to do so the authors have made the data available at https://wwwg2b.com/. (Summary by Rose McNelly @Rose_McN) Nature 10.1038/s41586-024-07682-9