Upstream ORFs check undesired leaky protein translation and buffer noise (Nature Plants)
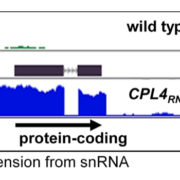
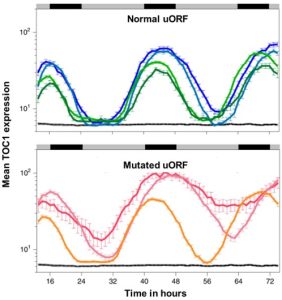 During my research journey, I have come across a handful of genes that are hard to express. Sometimes, even with a strong promoter and high transcript level, the protein level is negligible. We generally considered these proteins might be degrading very fast. But maybe not. A recent article by Wu et al. indicates translation of genes could be controlled by an upstream open reading frame (uORF). uORFs are upstream elements in the transcript that regulate the translation of about 30-50% of genes. The authors found that genes lacking uORF show leaky protein expression but the genes that have uORF don’t show this leaky expression. The authors observed the presence of uORF in a circadian-regulated gene TOC1. They mutated the uORF of TOC1 and observed that the mutated version produced more protein per mRNA than TOC1 harboring the normal uORF, indicating this uORF reduces protein expression. The authors also observed that the lack of proper uORF in Arabidopsis cells leads to loss of rhythmicity of TOC1 protein level, and that the uORF decreases cell-to-cell variability in TOC1 rhythmicity; that by buffering the expression level, noise is also reduced. They created a delightful animation that explains how this works. Overall, uORF elements seem to check undesired protein expression. Looking at the transcript of my hard-to-expressed genes, at least one has clear uORFs that might have limited the translation. (Summary by Kamal Kumar Malukani, @KamalMalukani) Nature Plants 10.1038/s41477-022-01136-8
During my research journey, I have come across a handful of genes that are hard to express. Sometimes, even with a strong promoter and high transcript level, the protein level is negligible. We generally considered these proteins might be degrading very fast. But maybe not. A recent article by Wu et al. indicates translation of genes could be controlled by an upstream open reading frame (uORF). uORFs are upstream elements in the transcript that regulate the translation of about 30-50% of genes. The authors found that genes lacking uORF show leaky protein expression but the genes that have uORF don’t show this leaky expression. The authors observed the presence of uORF in a circadian-regulated gene TOC1. They mutated the uORF of TOC1 and observed that the mutated version produced more protein per mRNA than TOC1 harboring the normal uORF, indicating this uORF reduces protein expression. The authors also observed that the lack of proper uORF in Arabidopsis cells leads to loss of rhythmicity of TOC1 protein level, and that the uORF decreases cell-to-cell variability in TOC1 rhythmicity; that by buffering the expression level, noise is also reduced. They created a delightful animation that explains how this works. Overall, uORF elements seem to check undesired protein expression. Looking at the transcript of my hard-to-expressed genes, at least one has clear uORFs that might have limited the translation. (Summary by Kamal Kumar Malukani, @KamalMalukani) Nature Plants 10.1038/s41477-022-01136-8