Twist of Fate: Ribosomal Stress Reprograms Root Hair Pattering
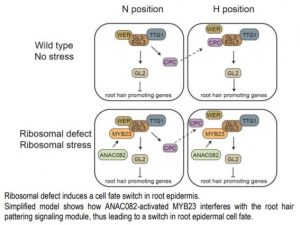
The root epidermis presents an elegant model to study cell differentiation. Based on positional cues, Arabidopsis distinguishes two epidermal cell types. Cells in the H position, adjacent to the junction of two cortex cells, have the capacity of developing root hair identity, whereas the non-hair cells occupy the N positions and are in contact with a single cortical cell. The fate of root epidermal cells lies in the hands of a complex of three transcription factors (TFs), WEREWOLF(WER), GLABRA 3/ENHANCER OF GLABRA 3 (GL3/EGL3), and TRANSPARENT TESTA GLABRA 1 (TTG1). In N position cells, this trio activates GLABRA 2 (GL2) and another TF, CAPRICE (CPC). GL2 represses transcription of a suite of root hair promoting genes, while the CPC protein is exported to an H position cell, where it counteracts WER function and subsequently represses GL2 (see figure). Thus, in the N position, cells with high GL2 expression inhibit root hair identity, while H position cells low GL2 expression activates root hair identity (Salazar-Henao et al., 2016)
 In the wer-1 mutant, N position cells gain root hair fate and most epidermal cells form a root hair. The cpc-1 mutant produces ectopic non-hair cells, but nevertheless around one third of its epidermis cells forms root hairs, suggesting additional regulators of root epidermis specification are yet to be discovered (reviewed in Salazar-Henao et al., 2016). Wang et al. (2020) addressed this issue with a cpc-1 enhancer screen. By analyzing an EMS-mutagenized population of the cpc-1 mutant, the authors identified a double mutant completely deprived of root hairs. The additional mutation was mapped to ARABIDOPSIS PUMILIO 23 (APUM23) which encodes a ribosome biogenesis factor. This suggests that a ribosomal defect can switch hair cell fate to a non-hair fate.
In the wer-1 mutant, N position cells gain root hair fate and most epidermal cells form a root hair. The cpc-1 mutant produces ectopic non-hair cells, but nevertheless around one third of its epidermis cells forms root hairs, suggesting additional regulators of root epidermis specification are yet to be discovered (reviewed in Salazar-Henao et al., 2016). Wang et al. (2020) addressed this issue with a cpc-1 enhancer screen. By analyzing an EMS-mutagenized population of the cpc-1 mutant, the authors identified a double mutant completely deprived of root hairs. The additional mutation was mapped to ARABIDOPSIS PUMILIO 23 (APUM23) which encodes a ribosome biogenesis factor. This suggests that a ribosomal defect can switch hair cell fate to a non-hair fate.
In order to dissect the mechanism of defective root pattering caused by ribosomal perturbation, Wang et al. (2020) examined expression of GL2 in the apum23-4 epidermis. While in wild type plants, GL2 expression is restricted to N position cells, lack of functional APUM23 resulted in GL2 transcription in some H cells as well. Next, the group examined GL2 expression in a series of crosses of apum23-4 plants with mutants disrupted in GL2 regulators. Mutations in GL3/EGL3 or TTG1 abolished GL2 transcription and the gl2‑1 apum23‑4 mutant developed no root hairs, whereas in the wer-1 apum23-4 double mutant GL2 was expressed in some epidermal cells and root hairs were formed in both N and H positions. These experiments provided the genetic evidence that a switch from hair to non-hair cell fate in the apum23‑4 mutant results from ectopic GL2 expression, which requires both GL3/EGL3 and TTG1, but is independent from WER. This finding opens up an avenue for elucidation of previously unidentified regulators of root epidermal cell patterning.
Since WER contribution to GL2 upregulation was excluded, the authors further searched for the TF that would act together with GL3/EGL3 and TTG1. MYB23, known previously to function redundantly to WER in N cells, appeared a likely suspect (Kang et al., 2009). Concomitant disruption of MYB23 and WER in apum23-4 mutant resulted in no GL2 expression. In wild type plants, MYB23 expression is confined to N position cells, but in apum23-4 roots was also observed in H position cells. MYB23 is also activated by WER-GL3/EGL3-TTG1 complex (Kang et al., 2009), yet the wer-1 mutation did not affect MYB23 expression in the apum23-4 line. In searching for a potential MYB23 activator, the authors used the anac082-1 mutant, previously described to reverse the developmental defects caused by ribosomal perturbations (Ohbayashi et al., 2017). Indeed, knocking out ANAC082 in apum23-4 mutant restored wild type root hair pattering, indicating that ANAC082 drives the MYB23‑dependent GL2 expression (Wang et al., 2020).
Wang and collaborators (2020) investigated how stress, expressed as altering ribosome biogenesis, plays a role in root epidermis specification. Ultimately the team demonstrated that ribosomal defects activate, independently from positional cues, an ANAC082-MYB23 signaling module, which by upregulation of GL2 leads to ectopic non-hair cell fate (see figure). Further studies may explain whether modulation of root hair development by environmental stresses, like salinity or nutrient deficiencies, and by ribosomal stress share any of these regulators. Identification of the apum23-4 mutant created an excellent system of perturbed root epidermis cell fate to elucidate the integration of stress responses and developmental regulation.
Dorota Kawa
Department of Plant Biology and Genome Center
University of California, Davis
ORCID: 0000-0002-4227-1621
REFERENCES
Kang, Y.H., Kirik, V., Hulskamp, M., Nam, K.H., Hagely, K., Lee, M.M., and Schiefelbein, J. (2009). The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21, 1080-1094.
Ohbayashi, I., Lin, C.Y., Shinohara, N., Matsumura, Y., Machida, Y., Horiguchi, G., Tsukaya, H., and Sugiyama, M. (2017). Evidence for a Role of ANAC082 as a Ribosomal Stress Response Mediator Leading to Growth Defects and Developmental Alterations in Arabidopsis. Plant Cell 29, 2644-2660.
Salazar-Henao, J.E., Velez-Bermudez, I.C., and Schmidt, W. (2016). The regulation and plasticity of root hair patterning and morphogenesis. Development 143, 1848-1858.
Wang, W., Ryu, K.H., Bruex, A., Barron, C., Schiefelbein, J. (2020). Molecular basis for a cell fate switch in response to impaired ribosome biogenesis in the Arabidopsis root epidermis. Plant Cell DOI: https://doi.org/10.1105/tpc.19.00773.



