Transcriptome landscape of a bacterial pathogen under plant immunity (PNAS)
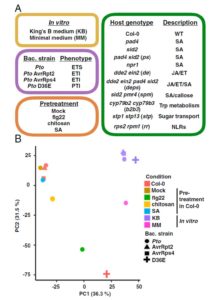 Many studies have examined how plants respond transcriptionally to pathogen attack. This study investigates how a bacterial pathogen [Pseudomonas syringae pv. tomato DC3000 (Pto)] alters its transcriptome very early in the infection process. To accomplish this, Nobori et al. developed two methods to enrich for rare bacterial transcripts against a much larger number of plant transcripts; in the first they isolated bacterial cells from infected plants prior to RNA isolation, and in the second they used a more expensive method involving removal of abundant plant RNAs with customized probes. Interestingly, these two methods led to very similar results. The authors looked at a single time point (6 hours post infection) but in a total of 27 combinations of plant and bacterial genotypes (including both plant and bacterial mutants affecting virulence or defense). In addition to generating rich data resources, this study also revealed some unexpected findings such as the possibility that the host manipulates the pathogen’s perception of iron as an antibacterial strategy. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1800529115
Many studies have examined how plants respond transcriptionally to pathogen attack. This study investigates how a bacterial pathogen [Pseudomonas syringae pv. tomato DC3000 (Pto)] alters its transcriptome very early in the infection process. To accomplish this, Nobori et al. developed two methods to enrich for rare bacterial transcripts against a much larger number of plant transcripts; in the first they isolated bacterial cells from infected plants prior to RNA isolation, and in the second they used a more expensive method involving removal of abundant plant RNAs with customized probes. Interestingly, these two methods led to very similar results. The authors looked at a single time point (6 hours post infection) but in a total of 27 combinations of plant and bacterial genotypes (including both plant and bacterial mutants affecting virulence or defense). In addition to generating rich data resources, this study also revealed some unexpected findings such as the possibility that the host manipulates the pathogen’s perception of iron as an antibacterial strategy. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1800529115