The Rules of Attachment: REC8 Cohesin Connects Chromatin Architecture and Recombination Machinery in Meiosis
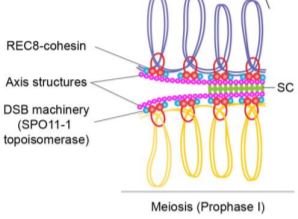
During cell division, accurate tethering and release of newly replicated sister chromatids is essential for ensuring that each daughter cell receives a full set of chromosomes. To accomplish this, cells use cohesins that act as the “chromosomal glue”. These cohesins are a group of ring-shaped protein complexes that are widely conserved from yeast to plants and humans. The core cohesin complex consists of four protein subunits, although individual subunits can vary to generate different forms with specialized functions. For instance, Sister Chromatid Cohesion 1 (SCC1) serves as the α-kleisin subunit of cohesins in somatic or mitotic cells. However, SCC1 is replaced by the α-kleisin RECOMBINATION 8 (REC8) specifically in meiotic cells.
The REC8-cohesin holds special importance in meiosis for its critical role in facilitating reciprocal DNA exchange between paired maternal and paternal chromosomes (i.e. crossover). Meiosis begins with the formation of hundreds of programmed DNA double-strand breaks (DSBs) on replicated chromatids. Most of these breaks are repaired by homologous recombination (using sister chromatids or homologous chromosomes as templates) without any reciprocal DNA exchange. Only a small fraction of meiotic DSBs (1- 4 per chromosome) are repaired to form crossovers. Defective meiotic cohesion and recombination in Arabidopsis rec8 mutants result in sterile gametophytes. How REC8 interacts with recombination machinery and other chromatin regulators to mediate chromosomal crossovers was incompletely understood.
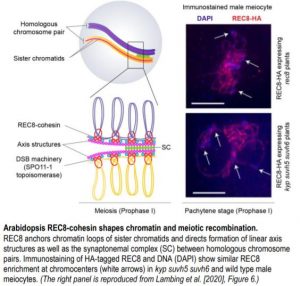 The recent study by Lambing et al. (2020) provides a comprehensive look into Arabidopsis REC8 (also known as SYNAPTIC1) localization patterns across meiotic chromosomes. The authors first generated Arabidopsis lines expressing hemagglutinin (HA)-tagged REC8 cohesins in the rec8 mutant background. REC8-HA plants rescue well-known rec8 mutant phenotypes such as impaired cohesion and abnormal axis structures between sister chromatids, and chromosome fragmentation. Next, the authors performed ChIP-Seq of unopened flowers from REC8-HA plants to identify genome-wide patterns of REC8 localization. To understand how REC8 influences meiotic chromatin and recombination, Lambing and colleagues assembled an impressive array of published datasets for nucleosome occupancy, DNA methylation, histone marks, recombination markers, and RNA expression. The authors found that REC8 is enriched in nucleosome-bound, repeat- and transposon-dense regions such as centromeres and pericentromeric heterochromatin. Repeat-rich regions rarely undergo meiotic recombination as these are highly susceptible to non-allelic crossovers that threaten genome stability (Choi et al., 2018). REC8 occupancy in such regions suggests that REC8 could suppress meiotic DSBs.
The recent study by Lambing et al. (2020) provides a comprehensive look into Arabidopsis REC8 (also known as SYNAPTIC1) localization patterns across meiotic chromosomes. The authors first generated Arabidopsis lines expressing hemagglutinin (HA)-tagged REC8 cohesins in the rec8 mutant background. REC8-HA plants rescue well-known rec8 mutant phenotypes such as impaired cohesion and abnormal axis structures between sister chromatids, and chromosome fragmentation. Next, the authors performed ChIP-Seq of unopened flowers from REC8-HA plants to identify genome-wide patterns of REC8 localization. To understand how REC8 influences meiotic chromatin and recombination, Lambing and colleagues assembled an impressive array of published datasets for nucleosome occupancy, DNA methylation, histone marks, recombination markers, and RNA expression. The authors found that REC8 is enriched in nucleosome-bound, repeat- and transposon-dense regions such as centromeres and pericentromeric heterochromatin. Repeat-rich regions rarely undergo meiotic recombination as these are highly susceptible to non-allelic crossovers that threaten genome stability (Choi et al., 2018). REC8 occupancy in such regions suggests that REC8 could suppress meiotic DSBs.
Lambing et al. (2020) examined this possibility by mapping REC8-enriched regions to previously reported meiotic DSB hotspots and crossover sites. REC8 and nucleosome colocalization indeed associates with meiotic DSB suppression. This pattern holds true even when the genomic landscape is limited to transposable element (TE)-containing regions. For instance, Arabidopsis TEs that move via RNA intermediates (RNA TEs) are known to be relatively more enriched in nucleosomes than TEs that move via DNA intermediates (DNA TEs). In agreement with this observation, RNA TEs also show REC8-enrichment and suppression of meiotic DSBs, while DNA TEs show the opposite trends. REC8 patterns thus corroborate the distinctive chromatin signatures associated with these two classes of transposons.
Arabidopsis TEs are transcriptionally repressed through heterochromatic marks such as cytosine DNA methylation and histone H3 lysine 9 dimethylation (H3K9me2). Since REC8 is enriched in heterochromatin, loss of the above marks could affect REC8 loading. Surprisingly, the authors found that REC8 recruitment to heterochromatin is generally unaffected even after drastic loss of H3k9me2 and non-CG DNA methylation in kyp suvh5 suvh6 triple mutants (see figure). However, a closer examination revealed reduced REC8 loading for a subset of RNA TEs belonging to the Copia superfamily. These Copia elements show increased expression and gain of meiotic DSB markers in kyp suvh5 suvh6 meiotic-stage flowers compared to that of wild type. By contrast, Copia elements with unchanged expression in kyp suvh5 suvh6 do not show any change in REC8 loading or meiotic DSB frequency. Thus, REC8 association with meiotic chromosomes is shaped by chromatin marks as well as transcription levels. Furthermore, REC8 mediates accurate polymerization of the axis proteins and the synaptonemal complex (SC), a protein scaffold that forms between homologous chromosomes during meiotic recombination. In rec8 mutants, abnormal axis structures can still form SC and recruit recombination machinery but to a much lesser extent than wild type.
Targeted meiotic recombination can enable increased genetic diversity and inheritance of desired traits in crops (Taagen et al. 2020). The work of Lambing et al. (2020) adds to the increasing knowledge on chromatin regulators that shape meiotic recombination in plants and paves the way for intimately dissecting the molecular processes governing this pathway.
Saima Shahid
Donald Danforth Plant Science Center
Saint Louis, Missouri
ORCID: 0000-0001-9385-0925
REFERENCES
Choi, K., et al. (2018). Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome research 28: 532–546.
Lambing, C. Tock, A.J., Topp, S.D., Choi, K., Kuo, P.C., Zhao, X. Osman, K., Higgins, J.D., Franklin, C.H., and Henderson, I.R. (2020). Interacting genomic landscapes of REC8-cohesin, chromatin and meiotic recombination in Arabidopsis thaliana. Plant Cell. https://doi.org/10.1105/tpc.19.00866
Taagen, E., Bogdanove, A. J., & Sorrells, M. E. (2020). Counting on crossovers: controlled recombination for plant breeding. Trends in Plant Science. https://doi.org/10.1016/j.tplants.2019.12.017