The NLR receptor ZAR1 has been guarding receptor-like cytoplasmic kinases since the Jurassic
Adachi et al. explore the conservation of a nucleotide-binding leucine-rich repeat immune receptor.
Hiroaki Adachi1,2,3, Toshiyuki Sakai1,2, Jiorgos Kourelis1, Hsuan Pai1, Jose L. Gonzalez Hernandez4, Yoshinori Utsumi5, Motoaki Seki5,6,7, Abbas Maqbool1 and Sophien Kamoun1*
1The Sainsbury Laboratory, University of East Anglia, Norwich Research Park, NR4 7UH, Norwich, UK
2Laboratory of Crop Evolution, Graduate School of Agriculture, Kyoto University, Mozume, Muko, Kyoto 617-0001, Japan
3JST-PRESTO, Saitama, Japan
4Agronomy, Horticulture and Plant Sciences Department, South Dakota State University, Brookings, South Dakota, US
5Plant Genomic Network Research Team, RIKEN Center for Sustainable Resource Science, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa 230-0045, Japan
6Plant Epigenome Regulation Laboratory, RIKEN Cluster for Pioneering Research, 2-1 Hirosawa, Wako, Saitama 351-0198, Japan
7Kihara Institute for Biological Research, Yokohama City University, 641-12 Maioka-cho, Totsuka-ku, Yokohama, Kanagawa 244-0813, Japan
Background: In plants, nucleotide-binding leucine-rich repeat (NLR) immune receptors generally exhibit hallmarks of rapid evolution even at the intraspecific level. NLRs evolve primarily through a birth-and-death process: new NLRs emerge by recurrent cycles of gene duplication and loss—some genes are maintained in the genome and acquire new pathogen detection specificities, whereas others are deleted or become non-functional through the accumulation of deleterious mutations. Such dynamic patterns of evolution enable the NLR immune system to keep up with fast-evolving effector repertoires of pathogenic microbes. Unlike typical NLRs, HOPZ-ACTIVATED RESISTANCE1 (ZAR1) is conserved across angiosperms.
Question: Can we use a molecular evolution framework to determine the critical features of a conserved plant NLR?
Findings: We performed iterative sequence similarity searches coupled with phylogenetic analyses to reconstruct the evolutionary history of ZAR1. ZAR1 is an atypically conserved NLR that traces its origin to early flowering plant lineages ~220 to 150 million years ago (Jurassic period). Ortholog sequence analyses revealed highly conserved features of ZAR1, including regions for pathogen recognition and immune activation. We functionally reconstructed the immune activity of ZAR1 and its host partner receptor-like cytoplasmic kinases (RLCKs) from distantly related plant species, supporting the hypothesis that ZAR1 has evolved to partner with RLCKs early in its evolution. ZAR1 stands out among angiosperm NLRs for having experienced relatively limited gene duplication and expansion throughout its deep evolutionary history.
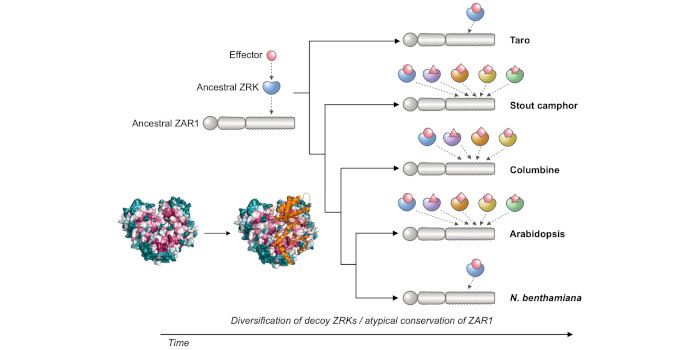
Next steps: Further comparative analyses, combining molecular evolution and structural biology, of plant and animal NLR systems will yield experimentally testable hypotheses for NLR research.
Reference:
Hiroaki Adachi, Toshiyuki Sakai, Jiorgos Kourelis, Hsuan Pai, Jose L. Gonzalez Hernandez, Yoshinori Utsumi, Motoaki Seki, Abbas Maqbool and Sophien Kamoun (2023) Jurassic NLR: conserved and dynamic evolutionary features of the atypically ancient immune receptor ZAR1. https://doi.org/10.1093/plcell/koad175