The 3′ processing of antisense RNAs physically links to chromatin-based transcriptional control (PNAS)
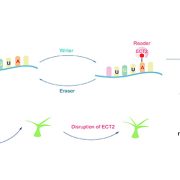
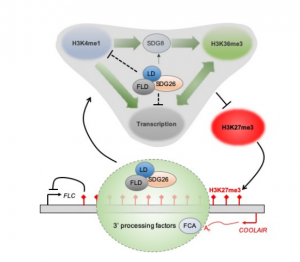 In Arabidopsis, multiple genetic pathways control the floral transition in response to external and internal stimuli. Among these, components of the autonomous pathway promote flowering by negatively regulating the central floral repressor FLOWERING LOCUS C (FLC). Despite the wealth of information on RNA-mediated chromatin silencing of FLC, the molecular link between FLOWERING TIME CONTROL (FCA)- a protein involved in RNA 3′ processing- and FLOWERING LOCUS D (FLD)- a demethylase involved in histone modification- is still missing. In this article, Fang and colleagues undertook a proteomic approach and found physical interaction between the chromatin modifiers FLD, LUMINIDEPENDENS (LD) and SET DOMAIN GROUP 26 (SDG26). Furthermore, Cross-Linked Nuclear Immuno-Precipitation and Mass Spectrometry (CLNIP–MS) analysis showed dynamic interaction of SDG26 not only with FLD-LD but also with different 3′ RNA processing factors including FCA. Additional genetic and molecular studies revealed increased levels of unspliced FLC RNA in higher order mutants, suggesting that the FLD-LD-SDG26 complex is recruited to repress FLC transcription upon RNA binding mediated by FCA. Lastly, quantitative detection of various histone modifications over the FLC gene body in loss of function mutants (fld, ld, sdg26, fca) indicated overaccumulation of histone marks related to transcriptional activation (H3K4me1, H3K36me3) and reduction of histone marks related to gene silencing (H3K27me3), causing late flowering. To conclude, the dynamic interactions between floral activators of the autonomous pathway result in a chromatin state associated with transcriptional inactivation of FLC. (Summary by Michela Osnato @Michela_Osnato) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2007268117
In Arabidopsis, multiple genetic pathways control the floral transition in response to external and internal stimuli. Among these, components of the autonomous pathway promote flowering by negatively regulating the central floral repressor FLOWERING LOCUS C (FLC). Despite the wealth of information on RNA-mediated chromatin silencing of FLC, the molecular link between FLOWERING TIME CONTROL (FCA)- a protein involved in RNA 3′ processing- and FLOWERING LOCUS D (FLD)- a demethylase involved in histone modification- is still missing. In this article, Fang and colleagues undertook a proteomic approach and found physical interaction between the chromatin modifiers FLD, LUMINIDEPENDENS (LD) and SET DOMAIN GROUP 26 (SDG26). Furthermore, Cross-Linked Nuclear Immuno-Precipitation and Mass Spectrometry (CLNIP–MS) analysis showed dynamic interaction of SDG26 not only with FLD-LD but also with different 3′ RNA processing factors including FCA. Additional genetic and molecular studies revealed increased levels of unspliced FLC RNA in higher order mutants, suggesting that the FLD-LD-SDG26 complex is recruited to repress FLC transcription upon RNA binding mediated by FCA. Lastly, quantitative detection of various histone modifications over the FLC gene body in loss of function mutants (fld, ld, sdg26, fca) indicated overaccumulation of histone marks related to transcriptional activation (H3K4me1, H3K36me3) and reduction of histone marks related to gene silencing (H3K27me3), causing late flowering. To conclude, the dynamic interactions between floral activators of the autonomous pathway result in a chromatin state associated with transcriptional inactivation of FLC. (Summary by Michela Osnato @Michela_Osnato) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2007268117